Figure 3.
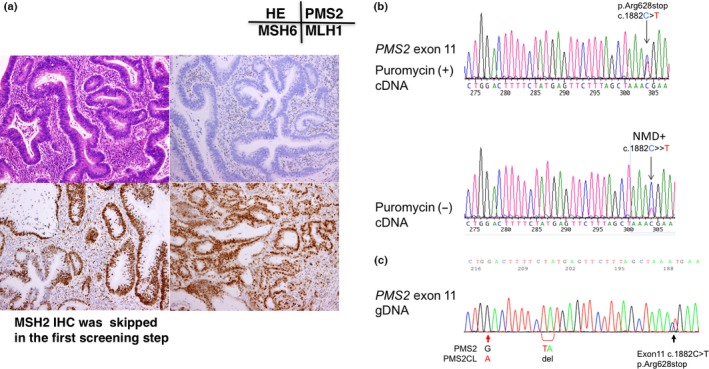
Immunochemical (IHC) and gene testing of subject No. 3. (a) IHC using four antibodies to mismatch repair (MMR) proteins. Note the loss of PMS2 protein expression in cancer cell nuclei. (b) RT‐PCR/direct sequencing analysis of the PMS2 gene in subject No. 3. The upper panel indicates the sequencing profile showing the c.1882C>T (p.Arg628Stop) mutation from the puromycin‐treated total RNA sample and the lower panel shows the sequencing profile without the puromycin treatment. Note that the signal ratio of the mutated allele (T) to the wild‐type allele (C) was weaker in the sample without the puromycin treatment than in that with the puromycin treatment, indicating nonsense‐mediated mRNA decay (NMD). (c) Long/nested PCR/direct sequencing analysis of the PMS2 gene. The black arrow indicates that the same mutation (c.1882C>T, pArg628Stop) was detected in the analysis using DNA by a long PCR/nested PCR/direct sequencing analysis. Red arrows indicate the nucleotide positions coding different DNA sequences between PMS2 and PMS2CL, showing that the sequences of the PMS2 paralogue were not contaminated in the PCR product.
