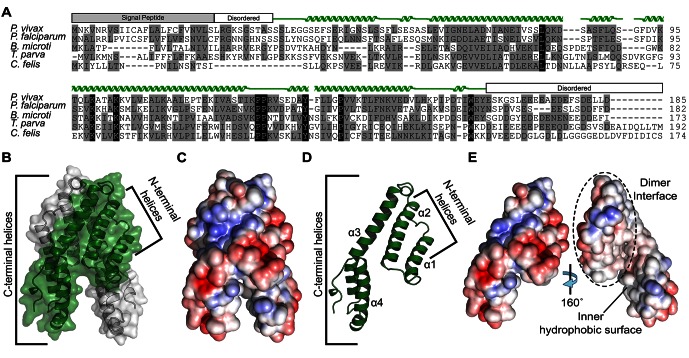
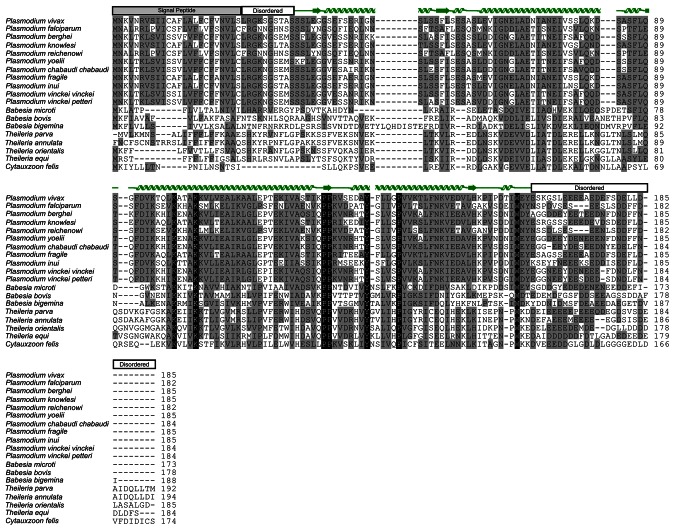
Figure 1. Alignment and structure of the conserved apicomplexan protein CelTOS from the human pathogen Plasmodium vivax (PvCelTOS).
(A) Alignment of CelTOS from apicomplexan parasites Plasmodium, Babesia, Theileria, Cytauxzoon, and mapping of the structural elements. In the alignment, grey shading represents similarity, black shading represents identity. The secondary structure features are based on the crystal structure of PvCelTOS and shown in green. Protein accession codes as follows: Plasmodium vivax [UniProtKB - A5JZX5], Plasmodium falciparum [UniProtKB - Q8I5P1], Babesia microti [UniProtKB - I7J9D8], Theileria parva [UniProtKB - Q4N982], Cytauxzoon felis [PiroplasmaDB - CF003135]. (B) PvCelTOS is an alpha helical dimer that resembles a tuning fork. Each monomer is in green or white ribbon and surface representation. (C) Surface maps of CelTOS dimer showing the electrostatic surface potential colored from red (−5 kT e−1) to blue (5 kT e−1). (D) The CelTOS monomer shown as ribbon representation can be separated into two distinct subdomains composed of N-terminal (α-helices 1 and 2) and C-terminal helices (α-helices 3 and 4). (E) Surface maps of CelTOS monomer reveals the inner hydrophobic surfaces composed of the dimer interface and one face of the tuning fork prongs. Coloring as in Figure 1C.
DOI: http://dx.doi.org/10.7554/eLife.20621.003