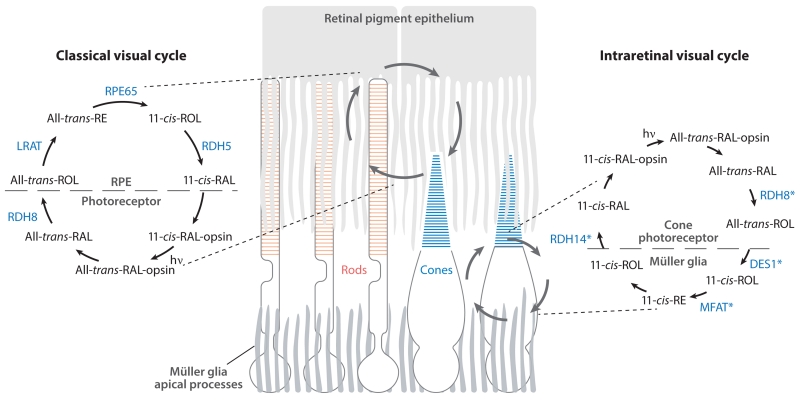
Figure 3.
Retinoid transformations and enzymes of the classical and intraretinal visual cycles. (Left) The classical visual cycle involving enzymes located in the RPE and photoreceptor outer segments. This pathway supports both rod and cone cell function. (Right) Reactions comprising the putative intraretinal visual cycle that provides cones with a privileged source of 11-cis-RAL. This pathway is thought to involve enzymes located in Müller glia and cone photoreceptors. Candidate enzymes of the pathway are shown in blue and marked by asterisks to indicate that their physiological involvement in the pathway has not yet been established. Abbreviations: DES1, dihydroceramide desaturase 1; hv, light; LRAT, lecithin:retinol acyltransferase; MFAT, multifunctional O-acyltransferase; RAL, retinal; RDH, retinol dehydrogenase; RE, retinyl ester; ROL, retinol; RPE, retinal pigment epithelium.