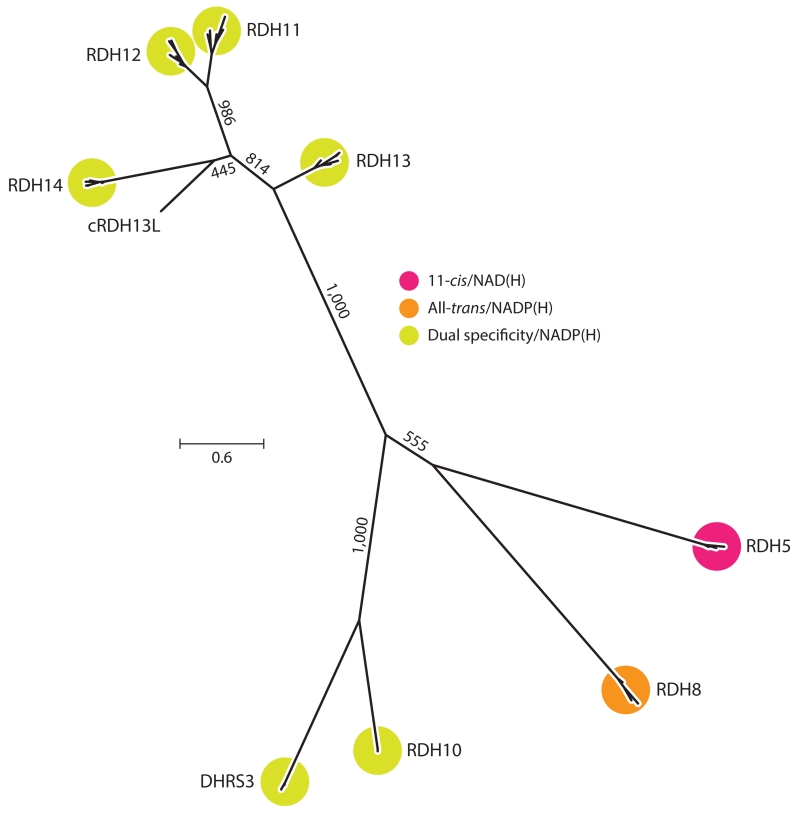
Figure 6.
An unrooted molecular phylogenetic tree illustrating relationships among groups of RDH homologs. Protein sequences (mammalian orthologs for each clade as well as carp RDH13L) were aligned with MUSCLE (Edgar 2004), and the tree was inferred using the phylogenetic maximum likelihood method as implemented in PhyML (ATGC Bioinformatics Platform, French National Institute of Bioinformatics, http://www.atgc-montpellier.fr/phyml/). Numbers on the internal branches indicate bootstrap support based on 1,000 pseudoreplicates. The scale bar indicates the average number of substitutions per site. The tree was generated using FigTree (Andrew Rambaut, http://tree.bio.ed.ac.uk/software/figtree/). Abbreviations: DHR, dehydrogenase/reductase family; NAD, nicotinamide adenine dinucleotide; RDH, retinol dehydrogenase.