Fig. 1.
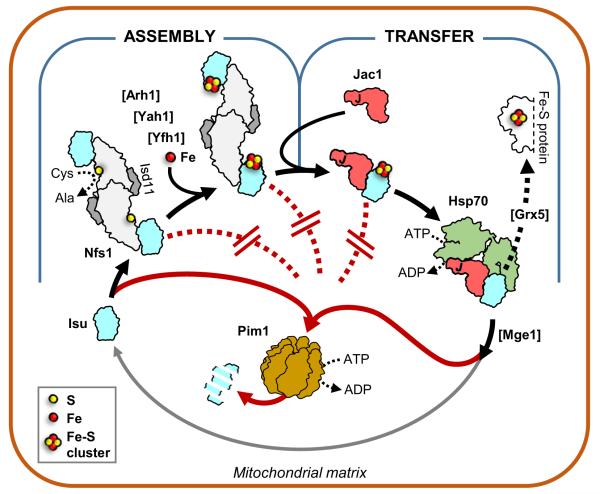
Mitochondrial Fe–S cluster biogenesis and Isu degradation. Model of regulation of Isu degradation in the mitochondrial matrix. Isu protein, a key component for Fe–S cluster biogenesis (black arrows), is susceptible to degradation by the Lon-type protease Pim1 (red arrows). However, Isu becomes protected from Pim1 driven degradation (red dotted lines) during both the assembly and transfer steps of the process through a combination of partner binding and Fe–S cluster coordination. Isu becomes susceptible to degradation upon completion of the transfer step. Assembly step. Fe–S cluster assembly is initiated by interaction of the scaffold Isu with Nfs1 (Lill et al. 2012). In the yeast S. cerevisiae the scaffold is encoded by two paralogous genes, ISU1 and ISU2; Isu1 and Isu2 are functionally redundant, with Isu1 being predominant and Isu2 less abundant (Schilke et al. 1999; Garland et al. 1999). As a cysteine desulfurase, Nfs1 provides sulfur for synthesis of the cluster. It functions as a Nfs1(Isd11) complex composed of two monomers of Nfs1 and two monomers of Isd11. Isd11 is an accessory protein required for Nfs1 stability and/or function (Wiedemann et al. 2006). Other factors needed for cluster assembly include: Yeast frataxin Yfh1, proposed to serve as an iron donor and/or regulator (Pastore and Puccio 2013), and yeast ferredoxin Yah1, which is functionally coupled with ferredoxin reductase Arh1, to provide electrons required for sulfur reduction (Webert et al. 2014). Transfer step. Following its assembly, the Fe–S cluster is transferred from Isu to a recipient protein (Lill et al. 2012). Transfer requires Hsp70 chaperone machinery composed of Hsp70, and its partnering co-chaperones Jac1 and Mge1 (Dutkiewicz et al. 2003). Initially Jac1 binds and targets the Isu coordinated Fe–S cluster to Hsp70. Interaction with Isu, coupled with Hsp70 ATPase activity stimulation by the J-domain of Jac1, results in Hsp70:Isu complex formation. As a result, the Fe–S cluster can be transferred via potential carrier proteins such as Grx5 (Uzarska et al. 2013) to the mitochondrial subset of recipient proteins or, through a yet elusive pathway, can be utilized for maturation of cytosolic and nuclear proteins (Lill et al. 2014). Isu is released from Hsp70, due to the nucleotide exchange activity of Mge1 protein (Dutkiewicz et al. 2003). Upon release, Isu becomes once more susceptible to degradation by Pim1, but can also be recycled and serve as a scaffold for cluster assembly again (gray arrow). Disruption at either the assembly or transfer step leads to increase in Isu levels as shown by analysis of yeast strains having reduced or perturbed function of Yah1, Yfh1, Jac1, Hsp70 or Grx5 protein (Andrew et al. 2008; Song et al. 2012)