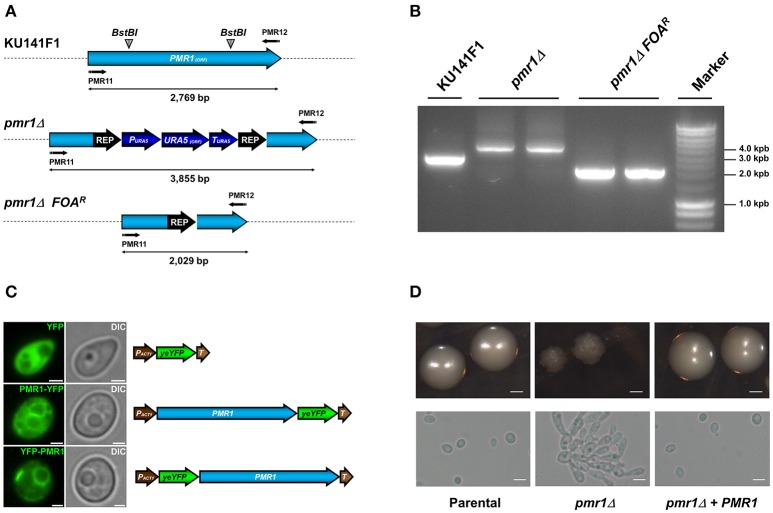
Figure 2.
Generation of the C. guilliermondii mutants used in this study and colony and cell morphology. (A) Schematic representation of the wild type and disrupted PMR1 loci. The PMR1 was disrupted in the strain KU141F1 using the URA5 blaster system. (B) Generation of the null mutant was confirmed by PCR reactions using the primer pair PMR11 5′-CTGAGAGGATCCATGACAGAGAACCCGTTCGATGTG-3′ and PMR12 5′-CTGAGAACTAGTTACGCTGTAGCTGACTCCATTCAT-3′ that amplifies the whole PMR1 locus. All the generated amplicons displayed the expected sizes, as indicated in (A) of the figure. (C) Subcellular localization of YFP alone, Pmr1-YFP and YFP-Pmr1 fusion proteins in the pmr1Δ strain. Scale bar = 2 μm. (D) Colony and cell morphology of C. guilliermondii strains. Cells were grown on SC medium at 30°C for 36 h, scale bar = 1 mm. Alternatively, strains were cultured in SC broth at 30°C for 18 h; scale bar = 10 μm. The strains used are: KU141 (Parental), HMY134 (pmr1Δ) and HMY138 (pmr1Δ + PMR1).