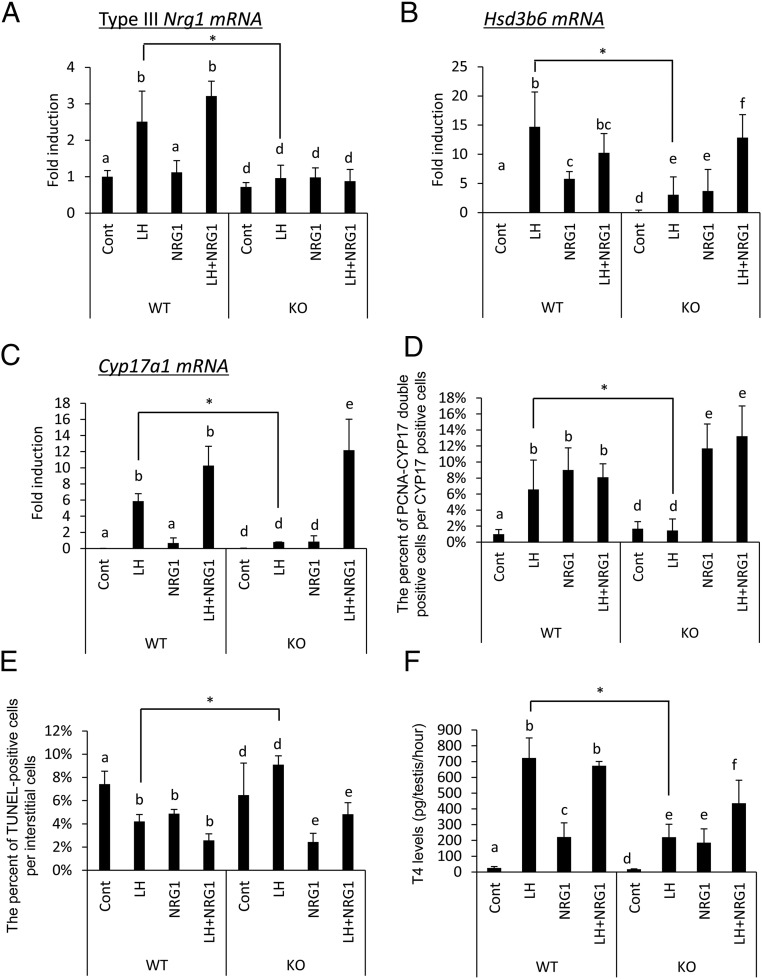
Figure 5.
NRG1 is essential for the proliferation of ALCs in infant testis cultured with LH. A–C, Testes from postnatal day 10 mice of both genotypes, WT and LeyNrg1KO (KO), were cultured for 3 days in media (control [cont]), LH (100 ng/μL), NRG1 (50 ng/μL), and LH+NRG1. The levels of mRNA encoding Nrg1 type III (a), Hsd3b6 (b), or Cyp17a1 (c) were analyzed by real-time PCR and normalized to that of L19. The value of controls (cont; no treatment) is set as 1. The data are presented as fold induction (mean ± SEM of three different experiments). The data were statistically analyzed by a two-way ANOVA. If a significant difference was observed in each factor, the comparison analysis was done by a Student's t test. Different superscripts denote significant differences among the treatment groups in each genotype (P < .05). *, Significant differences observed in each treatment group between genotypes. D and E, Testes from postnatal day 10 mice of both genotypes were cultured for 3 days with media alone (control [cont]), LH (100 ng/μL), NRG1 (50 ng/μL), and LH+NRG1. The number of PCNA-CYP17A1 double-positive (d) and TUNEL-positive (e) cells per section per each testis was analyzed (n = 3 for each treatment group and genotype). Values are presented as the mean ± SEM of three different cultured testes. The data were statistically analyzed by a two-way ANOVA. If the significant difference was observed in each factor, the comparison analysis was done by a Student's t test. Different superscripts denote significant differences among the treatment groups in each genotype (P < .05). *, Significant differences observed in each treatment group between genotypes. F, T (T4) levels in the culture medium after incubating testes from postnatal day 10 mice of both genotypes for 3 days with media alone (control [Cont]), LH (100 ng/μL), NRG1 (50 ng/μL), and LH+NRG1. The data were statistically analyzed by a two-way ANOVA. If the significant difference was observed in each factor, the comparison analysis was done by a Student's t test. Different superscripts denote significant differences among the treatment groups in each genotype (P < .05). *, Differences observed in each treatment group between genotypes.