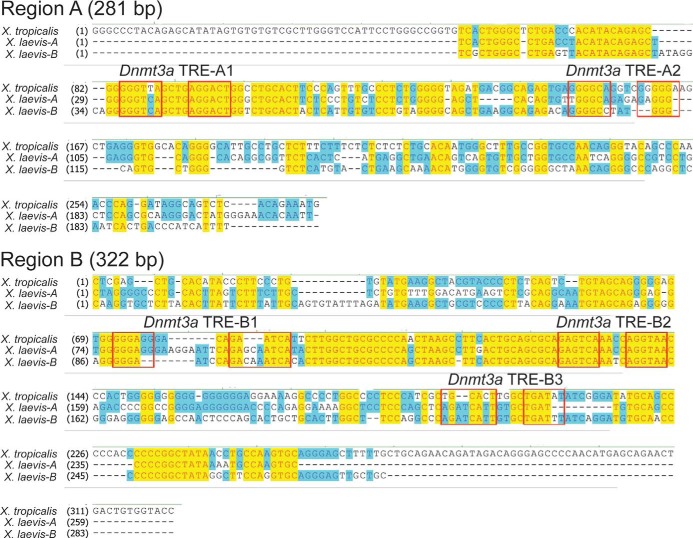
Figure 4.
Sequence alignment of regions A and B of X. tropicalis and X. laevis dnmt3a genes. Alignments were done using Clustal W. For X. tropicalis, we used genome build version 4.1, and for X. laevis we used genome build version 5.0 (there are two X. laevis genes owing to its pseudotetraploidy); region C is not shown because we found no evidence for TR association in brain chromatin by ChIP assay (Figure 2). Boxes indicate the locations of predicted DR+4 TRE half-sites numbered in the 5′ to 3′ direction based on the X. tropicalis reference genome. Note that the TRE-A1 and TRE-B2 sequences are conserved between the two frog species.