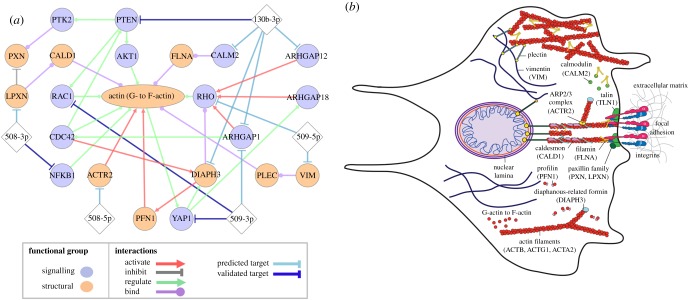
Figure 5.
Predicted effects of tumour-suppressor miRs on cytoskeletal structure and dynamics. (a) Network showing predicted and validated miR targets and functionally annotated protein–protein interactions for structural and signalling proteins that are known to regulate the mechanical properties of cells. Predicted miR–mRNA targets are determined using TargetScan v. 7.1, miRanda-mirSVR and miRmap; all targets displayed have scores in the top 50th percentile of predicted targets for all three methods, except for RAC1, which is a validated target of miR-509-3p [6]. Experimentally validated direct miR–mRNA targets are denoted by dark blue lines. Diamonds represent miRs; circles represent genes; and the ellipse represents actin, which exists as both monomers (G-actin) and filaments (F-actin). RHO denotes RhoA, RhoB and RhoC, which have high sequence homology [47–49]. While RHOC is a highly scoring predicted target of miR-509-5p, and RHOA is regulated by ARHGAP18 [50], the preferred substrates for other RhoGTPase-activating proteins are not fully understood. (b) Schematic illustrating regulation of the architecture and dynamics of the actin cytoskeleton.