Fig. 1.
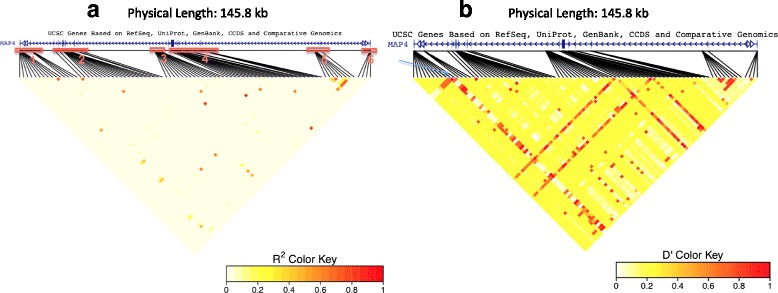
Pairwise LD measures for markers within MAP4 region on chromosome 3 in 1943 unrelated samples. The hg19 genome assembly was used for annotation. In panel (a), each pixel represents pairwise LD, measured by the squared allelic correlation coefficient r 2 between 2 markers. In panel (b), LD is measured by Lewontin’s |D '|. The latter are generally higher because |D '| takes into account that the correlation is constrained by the allele frequencies. As indicated by the color key, stronger LD is represented by red and weaker by white. The LD plot was produced using the LDheatmap package [16]
