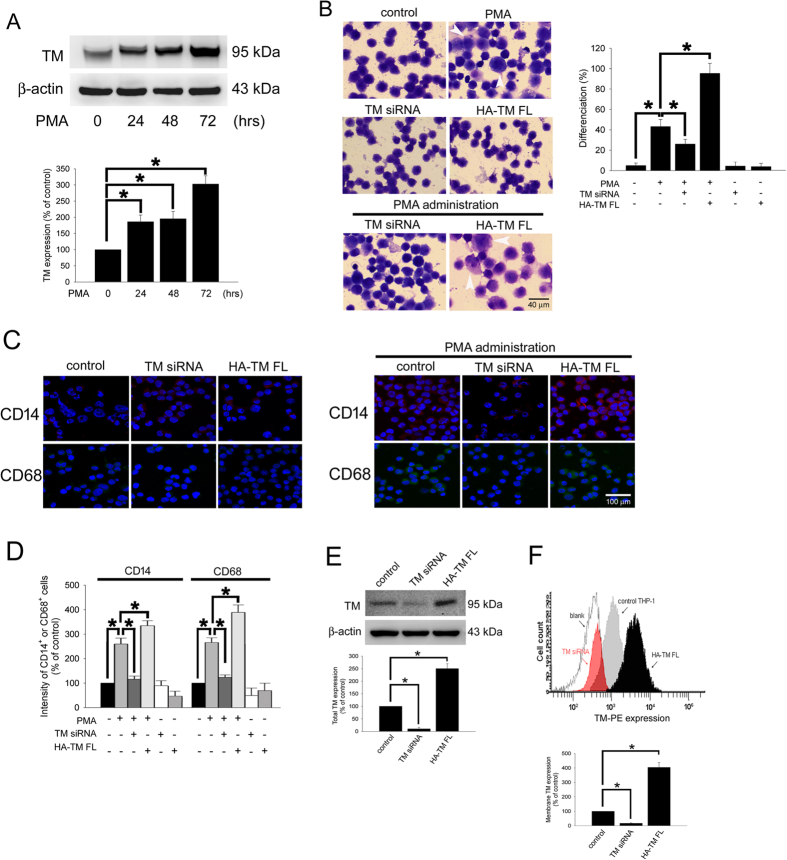
Figure 1. PMA-induced TM expression mediates morphological changes and differentiation marker expression in THP-1 cells.
(A) THP-1 cells were treated with 150 nM PMA for 24–72 hours. The total cell lysates were harvested, and the expression of TM was analyzed using western blot analysis. β-actin was used as a loading control. Five independent experiments have been performed (n = 5) and representative images have been showed. The amount of proteins expression was quantified using densitometry and presented as bar graph. The data are presented as the mean ± SD (n = 5), and *p < 0.05 was considered significant. (B) THP-1 cells were transfected with TM siRNA or HA-TM FL plasmid for 24 h followed by PMA stimulation for 72 hours. The morphology of the cells was observed using light microscopy. The adherent differentiated macrophage-like cells are indicated by a white arrowhead. Five independent experiments have been performed (n = 5). The quantification is shown in the right graph. (C) The expression of the macrophage cell surface markers CD14 (red) and CD68 (green) was analyzed using immunofluorescence and microscopy. Hoechst staining was used to label the nuclei. The scale bar indicates 100 μm. Five independent experiments have been performed (n = 5), and showed representative images. (D) The expression of CD14 and CD68 was analyzed using flow cytometry. Data are expressed as a % of the control, are presented as the mean ± SD and represent the results of three independent experiments (n = 3, *p < 0.05 was considered significant). (E) THP-1 cells were transfected with TM siRNA or HA-TM FL plasmid for 24 h, the TM in total cell lysate was analyzed using western blot analysis. The amount of proteins expression was quantified using densitometry and presented as bar graph. (F) The membrane TM was stained by PE-conjugated anti-TM antibody and analyzed using flow cytometry. The relative amount of membrane TM expression was presented as bar graph. The data are presented as the mean ± SD (n = 5), and *p < 0.05 was considered significant.