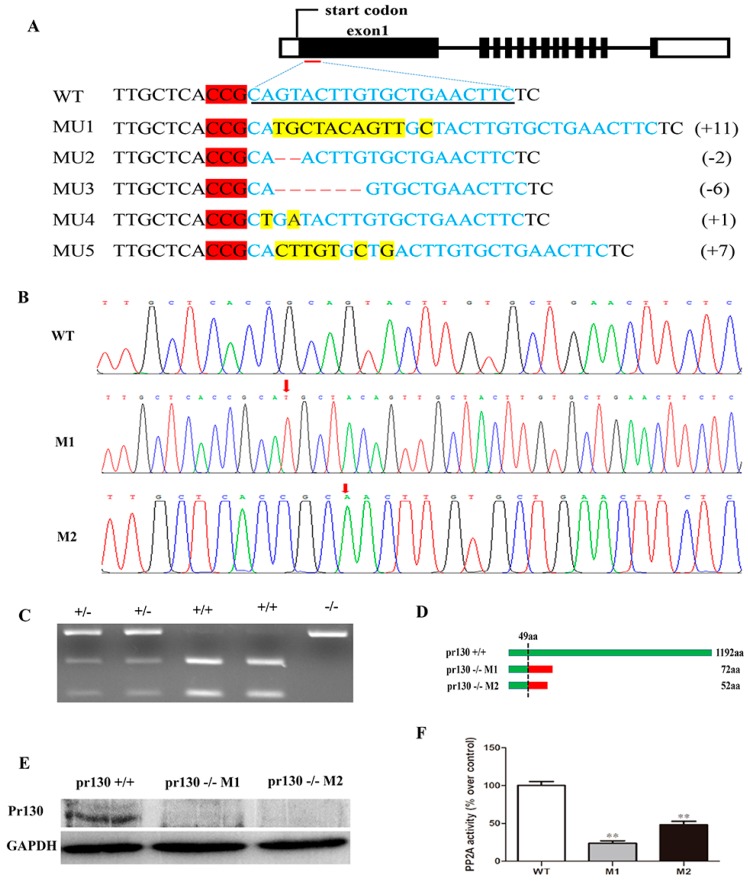
Figure 2.
Schematic diagram of pr130 knock-out in zebrafish using CRISPR/Cas9 system and analysis of PP2A activity. (A) Sequence of target site is underlined with black line and is shown in blue font in wild-type (WT) and protosacer adjacent motif (PAM) highlighted in red. Deletions and insertions are indicated by dashes and yellow highlight, respectively. Changed nucleotides are shown on the right side, “+” indicates insertion and “-” indicates deletion; (B) The sequences of WT, M1 and M2, the regions of insertion (M1) and deletion (M2) are indicated by red arrows; (C) A representative genotyping of ScaI digestion of PCR products amplified from genomic DNA, the products amplified from WT samples could be digested completely, whereas those of the homozygous mutants could not be digested, and those of the heterozygous mutants could be partially digested. +/+ indicates WT, (+/-) indicates heterozygous mutants, (-/-) indicates homozygous mutants; (D) The predicted amino acids of Pr130, in which showed a truncated protein, the first 49 aa (green) are identical to those of the WT Pr130 protein, which contains 23 (M1) or 3 (M2) miscoding amino acids (red); (E) Western blotting analysis of Pr130 protein; (F) PP2A activity was determined in cardiac extracts of WT and mutants. Data represent the mean ± SD. The symbols ** in the bar chart represent significant differences (p < 0.01).