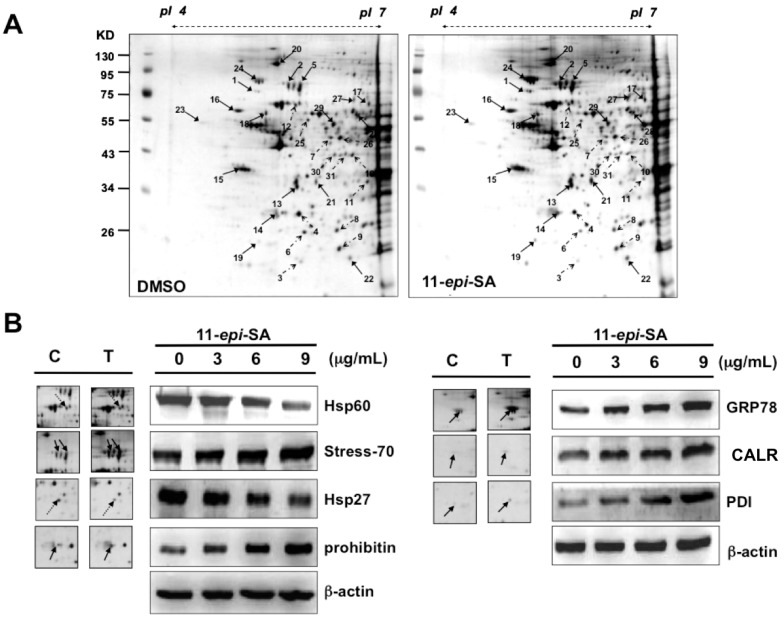
Figure 5.
Identification of differentially expressed proteins from mock- and 11-epi-SA-treated HA22T hepatoma cells by two-dimensional gel electrophoresis (2DE). (A) HA22T cells treated with DMSO (control) or 11-epi-SA at the concentration of 9 μg/mL for 24 h, were harvested and cell lysates were prepared as described in the Materials and Methods section. One hundred micrograms of protein were subjected to 2DE, and the proteins were visualized by silver staining. PDQuest image analysis software (Bio-Rad) was used for detecting the differential protein spots; (B) Validation of identified selected proteins from 2DE. The cell lysates prepared from mock- and 11-epi-SA-treated cells were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) for protein separation. Some identified proteins including glucose-regulated protein 78 (GRP78)/binding immunoglobulin protein (Bip), calreticulin (CALR), protein disulfide isomerase (PDI), 60 kDa heat shock protein (Hsp60), stress-70, 27 kDa heat shock protein (Hsp27), and prohibitin were detected by Western blotting by using specific antibodies as indicated. C: control, DMSO-treated cells; T: 11-epi-SA-treated cells. β-actin was used as the loading control.