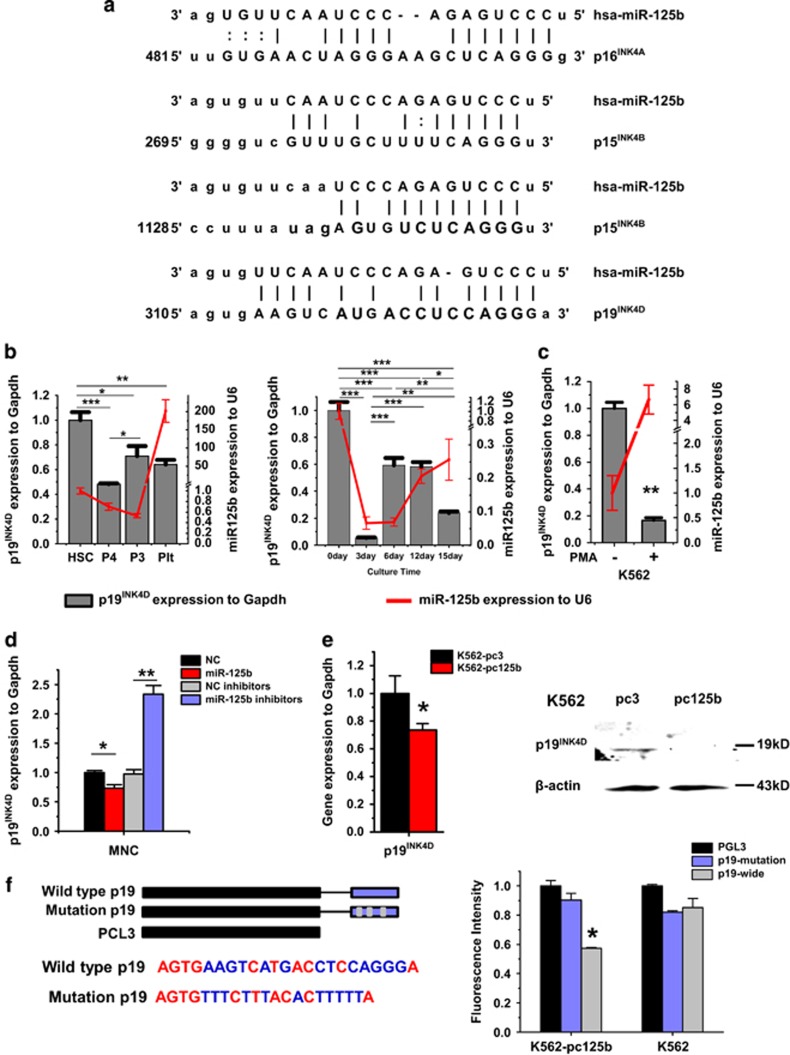
Figure 6.
Scan for miR-125b target genes during MK differentiation. P19INK4D is a direct target of miR-125b. (a) Bioinformatic analysis of the potential interactions between miR-125b and the 3′-UTR of genes from the INK4 family. (b) The expression pattern of p19INK4D during MK differentiation, as well as at distinct stages of maturation. The megakaryocytic induction begins with CD34+ hematopoietic precursors. (c) qPCR analysis of p19INK4D levels in untreated or PMA-treated K562 cells. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an endogenous expression control. (d) P19INK4D is upregulated upon miR-125b silencing; in contrast, p19INK4D is efficiently downregulated after miR-125b overexpression. (e) P19INK4D expression at the mRNA and protein levels in K562 cells stably transfected with pcDNA3.1 (control) or pcDNA3.1-pri-miR-125b were evaluated by qPCR and western blot, respectively. (f) Putative miR-125b binding sequence in the p19INK4D 3′-UTR, and the designed scheme for wild-type and mutant recognition fragments from p19INK4D 3′-UTR. Relative luciferase activity indicates direct binding and function of miR-125b on the p19INK4D 3′-UTR. miR-125b downregulates p19INK4D by interacting with its 3′-UTR. Relative repression of firefly luciferase activity was normalized to a transfection control. Individual comparisons between each groups were performed using Student's paired t-test. Bonferroni's multiple comparison test for multiple comparisons was applied. *P<0.05, **P<0.01 and ***P<0.001