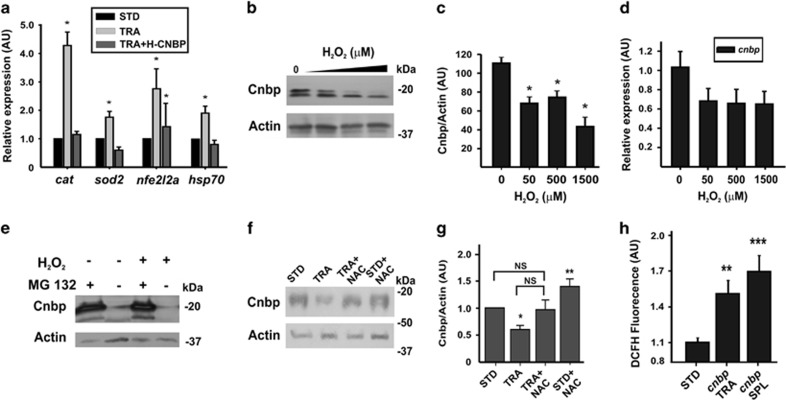
Figure 5.
Cnbp overexpression prevents the upregulation of oxidative stress-responsive genes. (a) Expression of stress-responsive genes: cat (encoding catalase); sod2 (encoding Mn-Superoxide Dismutase); nfe2l2a (enconding nuclear factor (erythroid-derived 2) like 2a) and hsp70 (encoding heat shock protein 70) in controls (STD), TRA and TRA+H-Cnbp embryos staged at 24 hpf (RT-qPCR). Bars represent mean of relative abundance±S.E.M., n=3, *P<0.05 versus STD, (t-test). (b) Western blotting detection of Cnbp in 24 hpf embryos exposed to different concentrations of H2O2. Actin was assessed as a loading control. Molecular weight markers are shown at the right. (c) Densitometric analysis of Cnbp normalized to Actin. The bars are means±S.E.M. of three experiments, *P<0.05 (ANOVA test). (d) Relative abundance of cnbp mRNA in samples from 24 hpf embryos exposed to different concentrations of H2O2, n=3. (e) Embryos at 30% epiboly were incubated in H2O2 or control conditions in the presence or the absence of the proteasome inhibitor MG-132. At 24 hpf, the abundance of Cnbp was assessed by western blotting. Actin is the loading control. Molecular weight markers are indicated to the right. (f) Western blotting showing the Cnbp abundance in control (STD), TRA, TRA+NAC and STD+NAC 24 hpf embryos. Molecular weight markers are shown at the right. Actin was analyzed as a loading control. (g) Densitometric quantification of western blotting bands (normalization of Cnbp signal with respect to Actin). Bars represent mean of relative abundance and S.E.M., n=3, *P<0.05; NS=non-significant P>0.05 (Mann–Whitney Test) (h) ROS accumulation in cnbp-knockdown 48 hpf embryos (cnbp TRA and cnbp SPL are embryos injected with Cnbp translation or splicing MO, respectively). Each value is expressed as the mean±S.E.M. (n=42). **P<0.01, ***P<0.001 (t-test). AU: arbitrary units