Fig. 5.
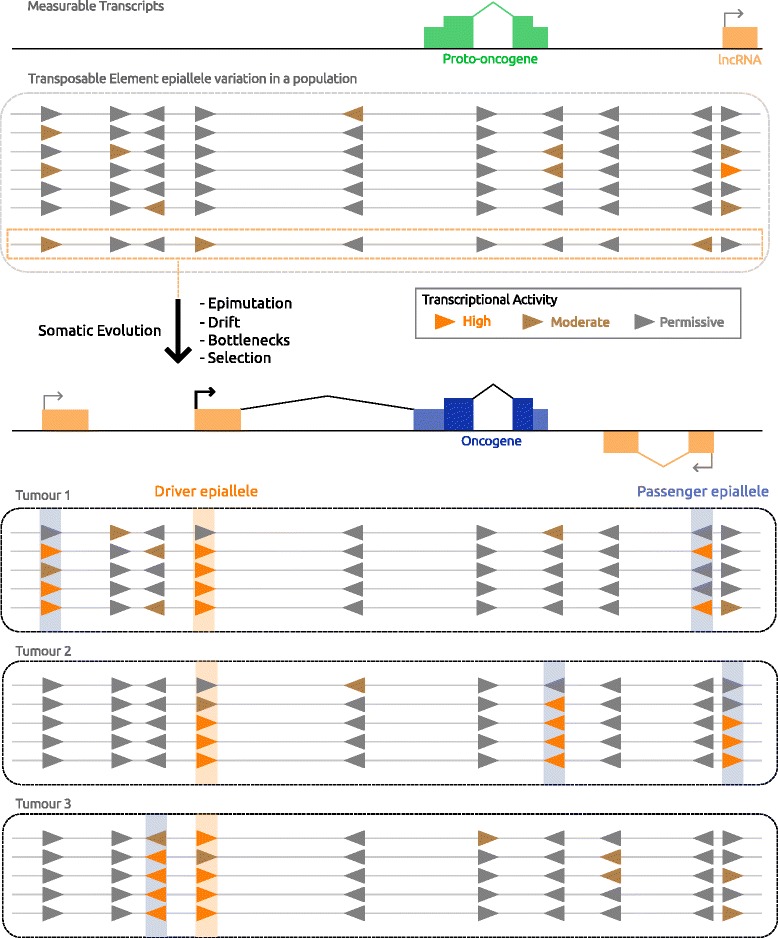
Epigenetic evolution model for onco-exaptation. In the starting cell population there is a dispersed and low/noisy promoter activity at TEs (colored triangles) from a set of transcriptionally permissive TEs (grey triangles). TE-derived transcript expression is low and variable between cells. Some transcripts are more reliably measurable (orange box). Clonal tumor evolutionary forces change the frequency and expression of TE-derived transcripts by homogenizing epialleles and use of TE promoters (highlighted haplotype). A higher frequency of ‘active’ TE epialleles at a locus results in increased measurable transcripts initiating from that position. TE epialleles that promote oncogenesis, namely onco-exaptations, can be selected for and arise multiple times independently as driver epialleles, in contrast to the more dispersed passenger epialleles, or “nonaptations”
