Figure 2. PAM recognition by Cse1 subunit of Cascade.
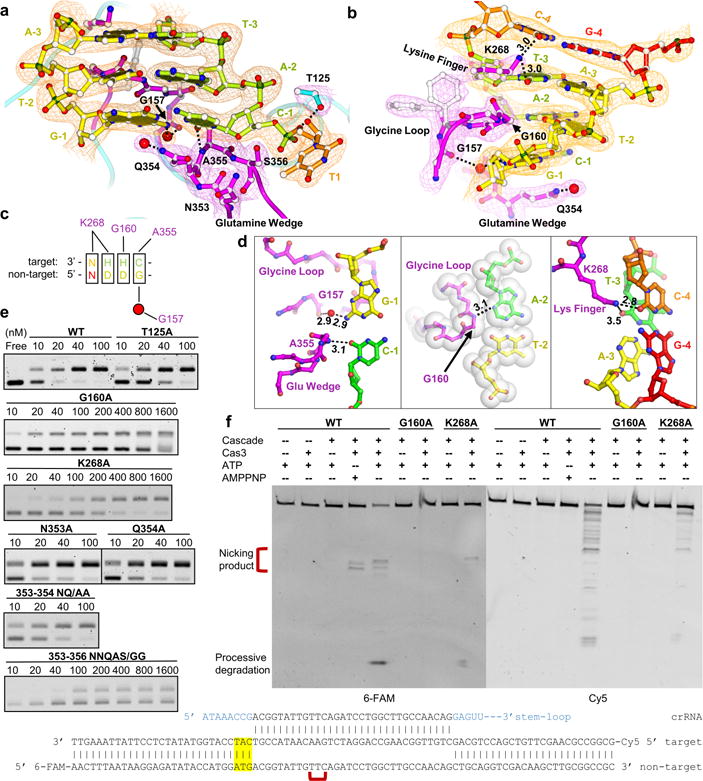
a, b, Detailed views featuring PAM recognition by the glutamine-wedge (and its involvement in target DNA strand displacement), glycine-loop, and lysine-finger of Cse1. The 2FO-FC electron density map is displayed at 1.5 σ. c, Summarization of the five interference PAMs in E. coli Type I-E CRISPR system, with the observed Cascade contacts marked. d, Top-down views of PAM recognition at each base-pair. Left: stringent recognition of PAM-1. Middle: shape complementarity to PAM-2. Right: Electrostatic contacts to PAM-3 and -4. e, Mutagenesis assayed by Cascade-binding (EMSA) and f, Cas3-mediated DNA cleavage to evaluate the observed PAM contacts.
