Figure 3.
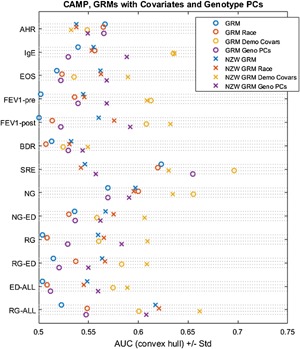
Prediction on CAMP cohort using GRMs with different covariates included, and a reduced set of Non‐Zero Weighted (NZW) SNPs. GRM, genetic relatedness matrix method using Leave‐One‐Out cross validation; AHR, airway hyperresponsiveness; EOS, eosinophil count; Pre‐FEV1, pre‐bronchodilator forced expiratory volume in 1 sec; Post‐FEV1, post‐bronchodilator forced expiratory volume in 1 sec; BDR, bronchodilator response ((Post‐FEV1 − Pre‐FEV1)/Pre‐FEV1); SRE, steroid responsiveness endophenotype; NG, normal FEV1 growth (without early decline); NG‐ED, normal FEV1 growth with early decline; RG, reduced FEV1 growth (without early decline); RG‐ED, reduced FEV1 growth with early decline; ED All, early FEV1 decline (with normal growth or with reduced growth); RG All, reduced FEV1 growth (with or without early decline). GRM‐only methods for IgE, EOS, post‐FEV1, NG, NG‐ED, RG, and RG‐All meet statistical significance for greater than random performance (AUC 0.50; p < 0.05, permutation test). Additionally, all combinations of the NZW GRM with clinical/demographic covariates were significant, except AHR and BDR.
