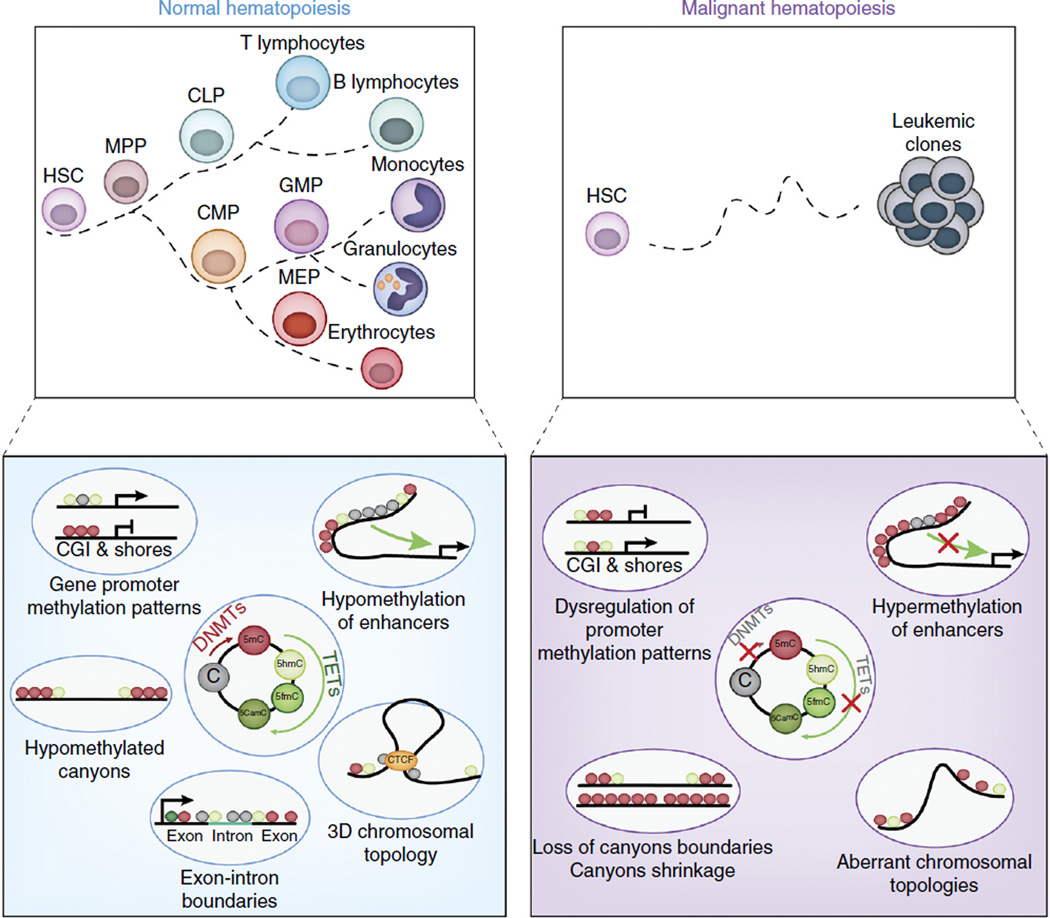
Figure 4.
Roles of the cytosine methylation and hydroxymethylation of DNA in normal and malignant hematopoiesis. The balance of the function of DNA methyltransferases (DNMTs) and TET enzymes is critical to the regulation of genome-wide localization and abundance of DNA cytosine modifications that in turn regulate gene-expression patterns required for normal hematopoiesis (top left). DNA methyltransferases catalyze the conversion of cytosine (C) into 5-methylcytosine (5mc), which serve as a substrate for the conversion of 5mC into 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fmC) and 5-carboxymethylcytosine (5cmC) by TET enzymes (bottom left). Loss or mutation of genes encoding DNA methyltransferases or TET enzymes, which commonly occur in a variety of leukemias, due to somatic mutations, perturbs the genome-wide distribution of 5mC and its oxidized derivatives (bottom right) and promotes leukemogenesis (top right). For example, hypermethylation might affect promoters or enhancers and result in gene silencing, or it might disrupt CTCF-binding sites to alter chromosomal structure and allow aberrant promoter-enhancer interactions to occur. Moreover, extended regions of low methylation that span domains containing transcription-factor-binding sites (so-called ‘canyons’) might shrink or widen with alterations in TET or DNMT function. MPP, multipotent progenitor; CLP, common lymphoid progenitor; CMP, common myeloid progenitor; GMP, granulocyte-macrophage progenitor; MEP, megakaryocytic-erythroid progenitor; CGI, CpG island.