Supplemental Digital Content is available in the text
Keywords: CTCs, EGFR mutation, NSCLC
Abstract
Background:
The value of circulating tumor cells (CTCs) in detecting epidermal growth factor receptor (EGFR) mutations in patients with nonsmall cell lung cancer (NSCLC) is controversial. We performed a meta-analysis to investigate the diagnostic significance of CTCs with tumor tissues as the standard control.
Methods:
A systematic literature search, including papers published until November 26, 2015, was performed using PubMed, Medline, Embase, Web of Science, and the China National Knowledge Infrastructure, and the references of retrieved articles were screened. The pooled sensitivity, specificity, and diagnostic odds ratio (DOR) were calculated according to the data selection from the included studies. The evaluation indexes of the diagnostic performance were the summary receiver operating characteristic curve (SROC) and area under the SROC (AUSROC).
Results:
Eight eligible articles with a total of 170 participants were identified in our meta-analysis. The pooled sensitivity and specificity were 0.91 [95% CI: 0.55–0.99] and 0.99 [95% CI: 0.59–1.00]. The positive likelihood ratio and negative likelihood ratio were 68 [95% CI: 1.4–3364] and 0.09 [95% CI: 0.01–0.64], respectively. The DOR was 788 [95% CI: 9–71884]. The high diagnostic performance of CTCs in detecting EGFR mutations was indicated by the AUSROC of 0.99 [95% CI: 0.98–1.00].
Conclusions:
CTCs are a feasible and highly specific biomarker for detecting the EGFR mutation status in NSCLC patients.
1. Introduction
Lung carcinoma is the leading cause of cancer-related mortality worldwide. There is an upward trend in the incidence of lung cancer with the widespread use of tobacco, tendency toward aging of the population and increased pollution of the environment. In China, this phenomenon is more obvious, as 1 institute showed a third of smokers in the world are Chinese, increasing the number of people who are exposed to smoke.[1] Moreover, 1 study reported that the relative risk is between 1.14 and 5.20 in people with second-hand or passive smoking exposure.[2] Even so, never smokers account for 25% of all lung cancer cases worldwide.[3] The overall 5-year survival for lung cancer is 16%,[4] which is largely because it is a lethal, late-stage diagnosis. Seventy percent of lung cancer diagnoses are in the late stage.[5] Thus, lung cancer is a great threat to anyone.
With the further development of medical research, individualized treatment for lung cancer has become possible, especially the emergence of targeted drugs. It is generally known that epidermal growth factor receptor (EGFR) mutations or gene amplification leads to hyperactivation of the EGFR signaling pathway in more than half of patients with nonsmall cell lung cancer (NSCLC),[6] accounting for 85% of lung cancer cases.[7] EGFR tyrosine kinase inhibitors (TKIs), such as gefitinib or erlotinib, could significantly prolong the progression-free survival and overall survival of NSCLC patients by inhibiting EGFR kinase activities.[8,9] However, the vast majority of EGFR-mutated patients invariably develop acquired resistance to TKIs.[10] Thus, the EGFR mutation status can directly influence the formulation of a therapeutic plan, affecting the survival of NSCLC patients. In other words, it is necessary to detect the EGFR mutation status of NSCLC patients.
Circulating tumor cells (CTCs) are cancer cells that are shed into the peripheral blood from primary tumors,[11] and they play a crucial role in tumor metastasis by traveling through the bloodstream to the organs.[12] CTCs exist in the peripheral blood of all major carcinomas, but they are not found in healthy subjects or patients with nonmalignant diseases.[13] Additionally, the American Society of Oncology first recommended that CTCs could be used as a tumor marker for clinical use in 2007.[14] Moreover, some reports demonstrated that CTC detection could confirm the tumor diagnosis[15–17]and have potential prognostic value in cancer.[18–20] Some studies have also suggested that the number of detected CTCs could be used to assess the response to chemotherapy.[21,22] CTCs were the originally recognized in liquid biopsy.[23] Currently, liquid biopsy analysis is becoming increasingly popular in translational cancer research because it is a noninvasive examination compared to tumor tissue, and it might be useful for dynamic observation of the diagnostic or therapeutic course in cancer patients.
At present, the value of CTCs in detecting the EGFR mutation status remains uncertain. In response, we conducted meta-analytic approaches to pool together the diagnostic accuracy of CTCs for detecting EGFR mutations compared with the “gold standard” of tumor tissues. Our findings provide a reference for future clinical work.
2. Materials and methods
2.1. Publication search
We performed an Internet search of PubMed, Medline, Embase, Web of Science, and the China National Knowledge Infrastructure, including all articles published from the inception of these databases until November 26, 2015, using the following terms: “lung cancer” or “lung neoplasms,” “epidermal growth factor receptor” or “EGFR,” “mutation,” “circulating tumor cells” or “CTCs.” There were no language restrictions. The references of all retrieved articles and those of relevant review articles were also manually screened. This study was planned, conducted, and reported in compliance with the standards of quality for reporting meta-analyses.[24]
2.2. Inclusion and exclusion criteria
Eligible studies had to meet all following inclusion criteria: NSCLC patients should be diagnosed by histopathology or cytology; the EGFR mutation status should be detected by CTCs and tumor tissues; and sufficient information for generating a diagnostic 2 × 2 table that includes true positives (TPs), true negatives (TNs), false positives (FPs), and false negatives (FNs). If the article mentioned that EGFR mutations exist in NSCLC and cancer in other patients was confirmed by histopathology or there were healthy subjects acting as the EGFR wild-type control, it was considered for inclusion.
The studies were excluded if they met any of the following criteria: the quantity of CTCs was insufficient to detect EGFR mutations; the EGFR mutation was not compared to tumor tissues; and there were duplicate data from the same center or insufficient information for generating the diagnostic 2 × 2 table.
2.3. Data extraction and quality scoring system
Two reviewers (YL and ZX) independently extracted the following data from all eligible studies: name of the first author, year of publication, the country in which the study was performed, blood sample volume; timing of blood withdrawal (pre-, during-, or post-treatment); method of CTC isolation and enrichment; techniques used for evaluating EGFR mutations in CTCs; EGFR mutation exon types; and number of TPs, FPs, TNs, and FNs. Disagreements were resolved by discussion with the third author (PZ).
The methodological quality of the studies were assessed using guidelines published by the standards for reporting diagnostic accuracy (STARD) initiative that aim to improve the quality of reporting in diagnostic studies.[25] The quality assessment of diagnostic accuracy studied (QUADAS) incorporated empirical evidence, expert opinion, and formal consensus to assess the quality of primary studies on the diagnostic accuracy.[26] The methodological qualities of the included studies were judged by the 2 reviewers (YL and ZX).
2.4. Statistical analysis
We selected the TP, FP, FN, and TN from the studies. The accuracy data were used to calculate the pooled sensitivity, specificity, negative likelihood ratio (NLR), positive likelihood ratio (PLR), diagnostic odds ratio (DOR), and 95% confidence intervals (95% CI). The combined sensitivity and specificity were measured by the DOR, which is computed as the PLR/NLR.[27] Additionally, the NLR is defined as (1-sensitivity)/specificity, while the PLR is defined as sensitivity/(1-specificity). The summary receiver operative curve (SROC) was generated and used to identify the sensitivity and specificity for the single test threshold in each study,[28] and the area under the SROC (AUSROC) was calculated.
The effect of the threshold was determined using the Spearman correlation between the logit of sensitivity and logit of 1-specificity, and a P value <0.05 indicated a significant threshold effect. The heterogeneity caused by the nonthreshold effect was measured using the Q test and inconsistency index (I2), and a P value ≤0.05 and I2 value ≥50% indicated significant heterogeneity caused by the nonthreshold effect. In the presence of significant heterogeneity, the DerSimonian–Laird method was used to calculate the estimates[29] and meta-regression was performed to detect the source. Deek funnel plot was using to detect the publication bias[30] and a P value <0.05 indicated the presence of publication bias.
All statistical analyses were performed using the STATA 12.0 package (StataCorp, College Station, TX) with the MIDAS module and Meta-Disc.
This study belongs to a meta-analysis, not need ethics committee certification at all.
3. Results
3.1. Study identification
As shown in the NSCLC flow chart (Fig. 1), a total of 106 potentially relevant published articles were sought. Of these, 28 remained after we scanned the abstracts. After further investigation 20 articles were removed (8 reviews, 5 articles about the detection method of CTCs, 4 articles about the value of prognosis and diagnosis, 1 article about CTCs used for EGFR expression, and 2 articles that did not provide sufficient data for constructing 2 × 2 tables). Finally, 8 studies[31–38] were included in this meta-analysis.
Figure 1.
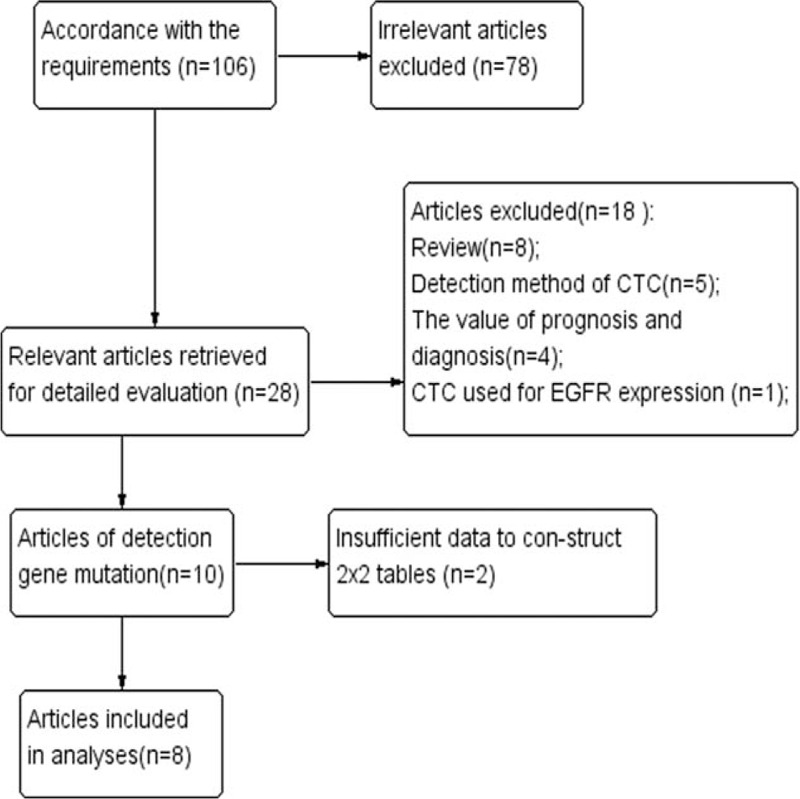
Flow diagram of study selection.
3.2. Characteristics of the eligible studies
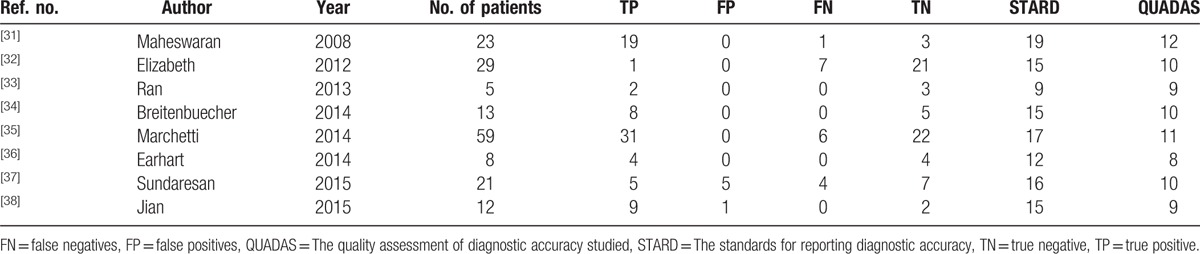
The clinical characteristics of these 8 eligible studies, including a total of 170 patients, are summarized in Table 1. One study incorporated patients from 9 study centers in the United States and Australia,[32] 3 studies evaluated patients from the United States, 2 from China, 1 from Italy, 1 from Germany, and 1 from the United States and Australia together. Most of the patients were NSCLC at the advanced or relapsed stage. The methods of CTC separation and gene status detection were not identical. The quality of the eligible studies and selection data for the TP, FP, FN, and TN are shown in Table 2. As shown, 6 studies had STARD scores ≥ 15[31,32,34,35,37,38] and 5 studies had QUADAS scores ≥ 10.[31,32,34,35,37]
Table 1.
Characteristics of eligible studies.
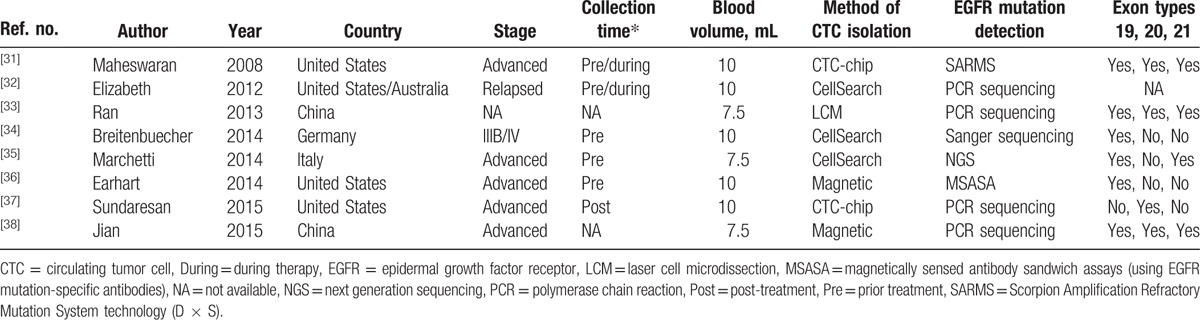
Table 2.
Summary data from the eligible studies.
3.3. Accuracy of CTCs for detecting EGFR mutations
Compared with tumor tissues in NSCLC patients, the pooled sensitivity and specificity of CTCs for detecting the EGFR mutation status were 0.91 [95% CI: 0.55–0.99] and 0.99 [95% CI: 0.59–1.00], respectively (Fig. 2). The PLR and NLR of CTCs were 68.04 [95% CI: 1.38–3364] and 0.09 [95% CI: 0.01–0.64], respectively (Fig. S1). The DOR was 788 [95% CI: 9–71884] (Fig. S2). As shown in Fig. 3, the AUSROC was 0.99 [95% CI: 0.98–1], indicating that CTCs had high diagnostic accuracy. Figure 4 contains a Fagan plot, which was generated for visual presentation of the diagnostic performance.
Figure 2.
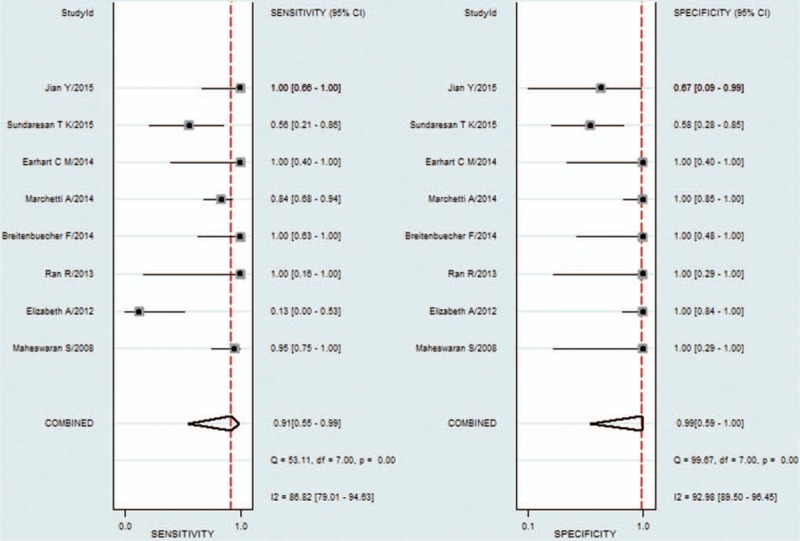
Forest plots of sensitivity and specificity of circulating tumor cells.
Figure 3.
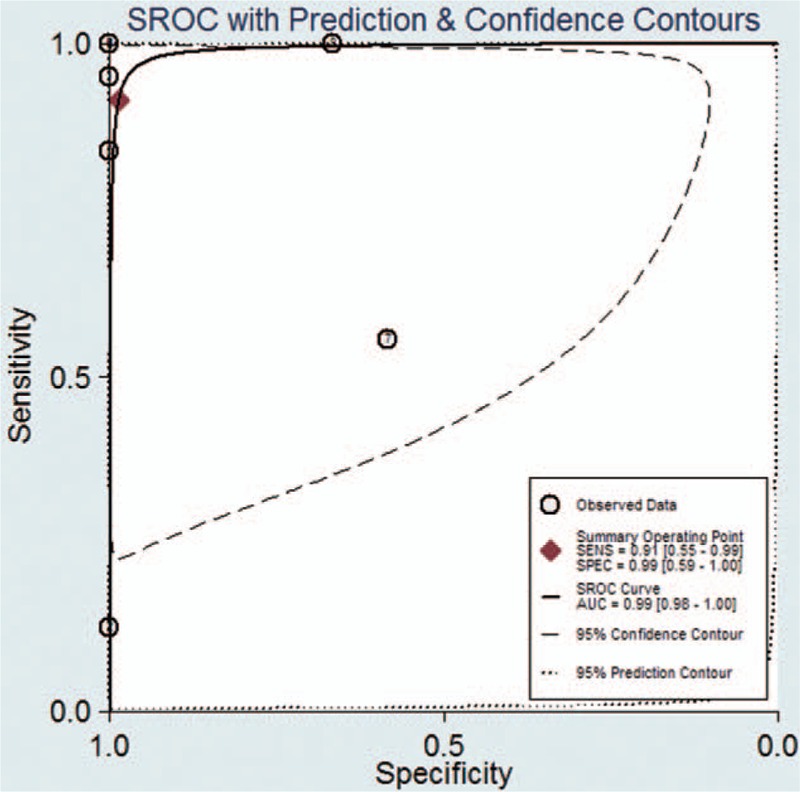
Area under the summary receiver operating characteristic curve.
Figure 4.
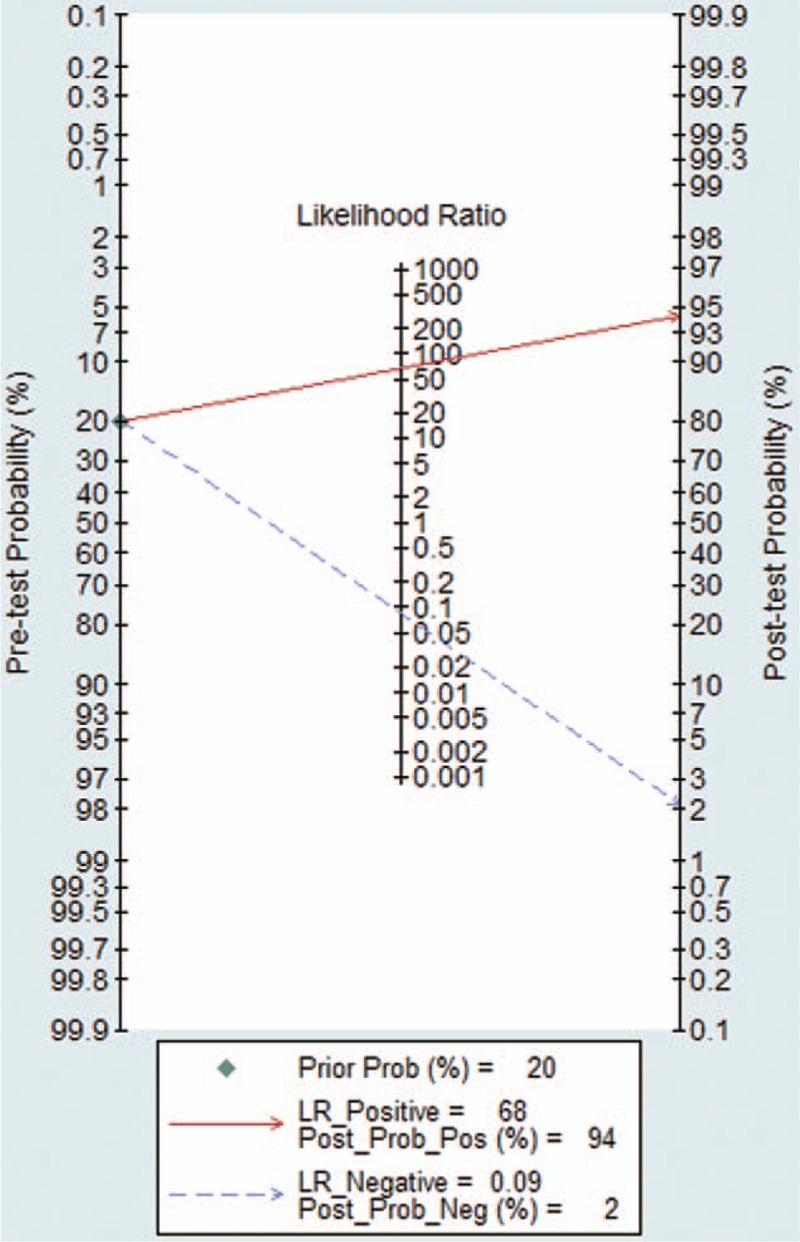
Fagan plot presents the clinical utility of circulating tumor cells.
3.4. Threshold effect and heterogeneity
One of the main sources of heterogeneity is the threshold effect. Visual assessment of the ROC plane suggested no significant threshold effect (Fig. S3). The threshold effect was calculated using the Spearman correlation coefficient and P value, which were 0.263 and 0.528 (>0.05), respectively, confirming that the threshold effect was not significant. As the forest plots of the accuracy data (sensitivity, specificity, PLR, NLR, and DOR) demonstrate, there was significant heterogeneity. Thus, multiple regression analysis was performed to test the source of heterogeneity and analyze the change in the diagnostic precision. However, these factors were not related to the heterogeneity.
3.5. Sensitivity analysis and publication bias
Deek funnel plot could respond to the publication bias. From the funnel plot (Fig. 5), there was no evidence of publication bias and the P value was 0.503 (>0.05), which also suggested the same result. In Fig. 6, we conducted a sensitivity analysis, and individual studies did not affect the pooled results.
Figure 5.
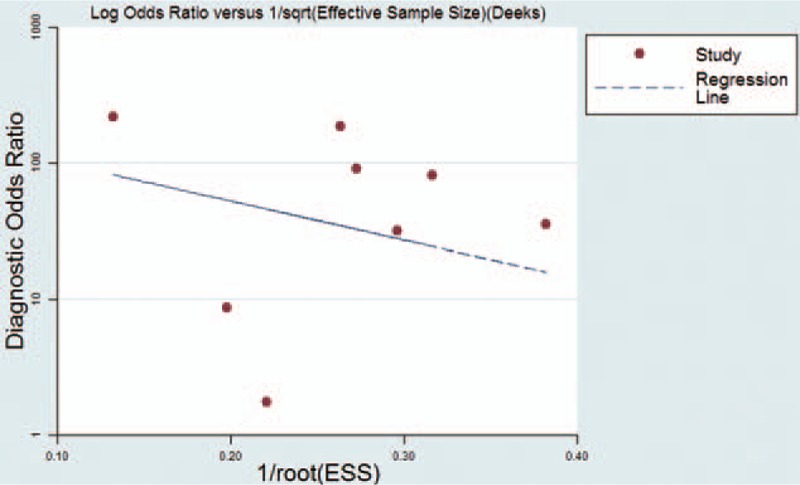
Deek funnel plot showed no significant publication bias.
Figure 6.
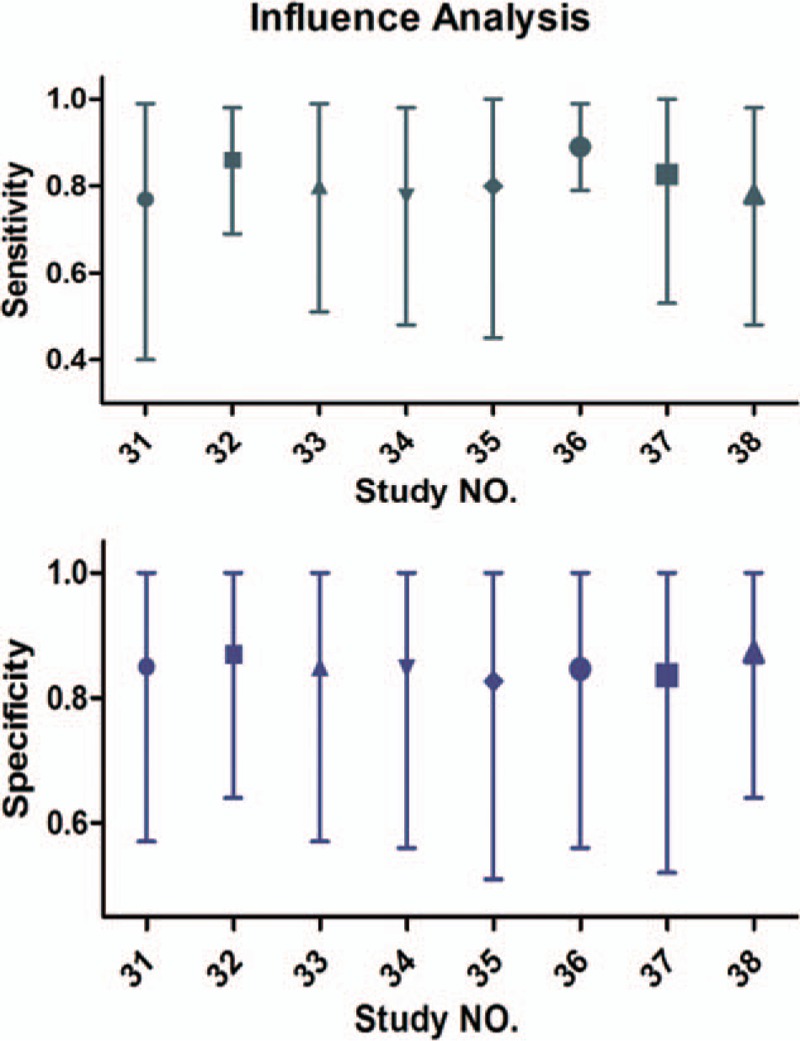
Sensitivity analysis showed that the pooled results were robust and not affected by individual studies.
4. Discussion
Evaluation of tumor tissues is the gold standard for diagnosing lung cancer, and this approach is used to detect the EGFR mutation status. However, it has numerous limitations. In the clinic, collecting multiple biopsies is not always feasible. As a result, a feasible, reliable, and minimally invasive approach for detecting EGFR mutations is needed. As a liquid biomarker, liquid biopsies conform to these requirements because they can easily be isolated from many body fluids (blood, saliva, urine, ascites, pleural effusion, etc.).[39] As a type of liquid biopsy, Ashworth first proposed the concept of CTCs in 1869,[11] and the presence of CTCs has been associated with a worse prognosis in several major cancers. As a more feasible and less invasive alternative, CTCs should be more widely used. In our meta-analysis, the pooled sensitivity and specificity for CTCs were 0.91 and 0.99, respectively. Additionally, the high AUSROC (0.99) indicated an overall high diagnostic accuracy of CTCs. Considering that a feasible, noninvasive biomarker with optimal performance is needed and the controversy over whether CTCs could provide a satisfying diagnostic accuracy, this meta-analysis offers important evidence to address the theory.
CTCs could detect both the common exon mutations of EGFR, such as 19-del, T790M, and L858R, and the rare exon types of EGFR (L861Q/A871G/L747_A750 del),[35] ALK,[40] KRAS, and BRAF.[32] However, combination analysis in EGFR and the ability to detect different exons of EGFR in CTCs is imperative. Among 40 cases with known EGFR exon 19 deletion mutations in primary tumors, 39 cases were positive in CTCs, suggesting that CTCs offer a highest diagnostic accuracy for EGFR exon 19 deletions. The detection rates of T790M and L858R were 60% (12/20) and 68.8% (11/16), respectively. Certainly, this is only based on part of the data because some studies did not report on the EGFR exon types or corresponding number.[31–33]
Improved lung cancer outcomes are inevitable with the advent of EGFR-TKI for EGFR mutations in lung cancer patients, but resistance to TKIs will eventually appear. The known mechanisms of drug resistance include T790M and PI3KCA mutations, MET amplification, HGF overexpression, epithelial to mesenchymal transition, BRCA1 mRNA expression, components of the NF-κB pathway and more.[41] Of these, more than a half of cases are attributable to the T790M mutation in exon 20 of EGFR.[42,43] AZD9291, as a novel, irreversible EGFR-TKI, has robust efficacy, and it is well tolerated in NSCLC patients with T790M mutations, resulting in acquired resistance to EGFR-TKIs based on a phase I clinical study.[44] In other words, resistance to EGFR-TKI based on a T790M mutation could be reversed to a certain extent. A T790M mutation is a secondary mutation that needs to be detected again. However, as we well know, only a few groups perform repeated tumor biopsies at the time of tumor progression. CTCs might overcome this difficulty as they offer a feasible, convenient and noninvasive approach. CTCs only detected the T790M mutation in 60% of samples in this meta-analysis, but substantial pieces of data were missing, and combining CTCs and circulating tumor DNA (ctDNA) could help identify the presence of T790M for patients in whom the concurrent biopsy was negative or indeterminate.[37]
There are many methods for detecting CTCs in the blood, and they belong to 2 main categories, namely, detection approaches and enrichment approaches. The former involves the identification portion, including cytometric and nucleic acid techniques, and the latter is geared toward isolation, including morphological and immunological techniques.[45] However, the efficiency for capturing CTCs is imparity between the different methods. As a result, the methods for CTC examination can affect detection of the EGFR mutation status. EGFR is one of the major oncogenes identified in a variety of human cancers, including breast cancer,[46] EGFR mutations are rare in human breast cancer.[47,48] In this meta-analysis, 2 studies[35,38] collected the blood samples from breast cancer patients without identifying EGFR mutations in their primary tumors or collected samples from healthy subjects as negative controls. We also adopted these 2 reports considering the situations described would not affect the outcome.
The heterogeneity of this meta-analysis should also be highlighted, but heterogeneity was not caused by the threshold effect of combining the results for the spearman correlation and ROC plane. The methods of isolation and detection of CTCs are varied[49] and the EGFR mutation statuses differ between the different races; these factors might lead to the observed heterogeneity. However, multivariable meta-regression showed that none of the analyzed covariates was the source of heterogeneity. It is unfortunate that we did not identify the source of heterogeneity. But, individual heterogeneity and tumor heterogeneity in the study may be the most important sources resulting in the heterogeneity.
Meta-analysis has been confirmed that ctDNA had high accuracy and specificity in the diagnosis of EGFR mutations.[50] Both CTCs and ctDNA have different characteristics, with same advantages such as minimally invasive, safe, convenient and it is acceptable by the patients. But, the primary advantage of CTCs over ctDNA is the period of emergence in the blood. CTCs are shed by primary and metastatic into the vasculature,[51] there could detect the CTCs prior to imaging among a handful of high-risk population of cancer; however, ctDNAs are released into the bloodstream following apoptosis or necrosis of cancer cells,[52] so, the accuracy of the CTC is superior to ctDNA in the early-stage NSCLC.
This is the first meta-analysis of the diagnostic performance of CTCs for detecting the EGFR mutation status. The results might provide guidance for future studies and clinical practice. Although the publication bias was not significant, there are several limitations. First, most studies were retrospective studies and the tissue samples were formalin-fixed paraffin-embedded, which leads to significant DNA degradation and increases the detection bias.[53] Second, only studies that were published in English and Chinese were included in our meta-analysis, which might create selection bias.[54] Third, eligible studies were lacking and the sample size of 170 participants was still relatively small, which would influence the statistical power.[55] As a result, a larger sample size and further research are needed.
5. Conclusions
In summary, the current evidence suggests a potential role for CTCs assays in confirming detection of the EGFR mutation status. The application range of CTCs in real-time monitoring of disease states in patients with NSCLC is very extensive due to its high specificity and noninvasive nature.
Supplementary Material
Footnotes
Abbreviations: AUSROC = area under the SROC, CTC = circulating tumor cell, DOR = diagnostic odds ratio, EGFR = epidermal growth factor receptor, NLR = negative likelihood ratio, NSCLC = nonsmall cell lung cancer, PLR = positive likelihood ratio, SROC = summary receiver operating characteristic curve.
This work was supported by grants from the National Natural Science Foundation of China (No. 81302032, 81401903, 81572937, and 81572273) and the Natural Science Foundation of Jiangsu province (No. BK2011658 and BK20140736).
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Zhang H, Cai B. The impact of tobacco on lung health in China. Respirology 2003;8:17–21. [DOI] [PubMed] [Google Scholar]
- [2].Ruano-Ravina A, Figueiras A, Barros-Dios JM. Lung cancer and related risk factors: an update of the literature. Public Health 2003;117:149–56. [DOI] [PubMed] [Google Scholar]
- [3].Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin 2005;55:74–108. [DOI] [PubMed] [Google Scholar]
- [4].McErlean A, Ginsberg MS. Epidemiology of lung cancer. Semin Roentgenol 2011;46:173–7. [DOI] [PubMed] [Google Scholar]
- [5].Yoder LH. Lung cancer epidemiology. Medsurg Nursing 2006;15:171–4. [PubMed] [Google Scholar]
- [6].Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol 2011;12:175–80. [DOI] [PubMed] [Google Scholar]
- [7].Cruz CSD, Tanoue LT, Matthay RA. Lung cancer: epidemiology, etiology, and prevention. Clin Chest Med 2011;32:605–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Costa DB, Kobayashi S, Tenen DG, et al. Pooled analysis of the prospective trials of gefitinib monotherapy for EGFR-mutant non-small cell lung cancers. Lung Cancer 2007;58:95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009;361:958–67. [DOI] [PubMed] [Google Scholar]
- [10].Hammerman PS, Jänne PA, Johnson BE. Resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res 2009;15:7502–9. [DOI] [PubMed] [Google Scholar]
- [11].Ashworth TR. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aust Med J 1869;14:146–9. [Google Scholar]
- [12].O’Flaherty JD, Gray S, Richard D, et al. Circulating tumour cells, their role in metastasis and their clinical utility in lung cancer. Lung Cancer 2012;76:19–25. [DOI] [PubMed] [Google Scholar]
- [13].Allard WJ, Matera J, Miller MC, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res 2004;10:6897–904. [DOI] [PubMed] [Google Scholar]
- [14].Harris L, Fritsche H, Mennel R, et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol 2007;25:5287–312. [DOI] [PubMed] [Google Scholar]
- [15].Msaouel P, Koutsilieris M. Diagnostic value of circulating tumor cell detection in bladder and urothelial cancer: systematic review and meta-analysis. BMC Cancer 2011;11:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004;351:781–91. [DOI] [PubMed] [Google Scholar]
- [17].Tanaka F, Yoneda K, Kondo N, et al. Circulating tumor cell as a diagnostic marker in primary lung cancer. Clin Cancer Res 2009;15:6980–6. [DOI] [PubMed] [Google Scholar]
- [18].Okabe H, Tsunoda S, Hosogi H, et al. Circulating tumor cells as an independent predictor of survival in advanced gastric cancer. Ann Surg Oncol 2015;22:3954–61. [DOI] [PubMed] [Google Scholar]
- [19].Krebs MG, Sloane R, Priest L, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol 2011;29:1556–63. [DOI] [PubMed] [Google Scholar]
- [20].Romiti A, Raffa S, Di Rocco R, et al. Circulating tumor cells count predicts survival in colorectal cancer patients. J Gastrointestin Liver Dis 2014;23:279–84. [DOI] [PubMed] [Google Scholar]
- [21].Okegawa T, Itaya N, Hara H, et al. Circulating tumor cells as a biomarker predictive of sensitivity to docetaxel chemotherapy in patients with castration-resistant prostate cancer. Anticancer Res 2014;34:6705–10. [PubMed] [Google Scholar]
- [22].Hirose T, Murata Y, Oki Y, et al. Relationship of circulating tumor cells to the effectiveness of cytotoxic chemotherapy in patients with metastatic non-small-cell lung cancer. Oncol Res 2012;20:131–7. [DOI] [PubMed] [Google Scholar]
- [23].Alix-Panabières C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem 2013;59:110–8. [DOI] [PubMed] [Google Scholar]
- [24].Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med 2009;151:264–9. [DOI] [PubMed] [Google Scholar]
- [25].Bossuyt PM, Reitsma JB, Bruns ED, et al. Towards complete and accurate reporting of studies of diagnostic accuracy: the STARD initiative. Clin Chem Lab Med 2003;41:68–73. [DOI] [PubMed] [Google Scholar]
- [26].Whiting P, Rutjes AWS, Reitsma JB, et al. The development of QUADAS: a tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med Res Methodol 2003;3:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Glas AS, Lijmer JG, Prins MH, et al. The diagnostic odds ratio: a single indicator of test performance. J Clin Epidemiol 2003;56:1129–35. [DOI] [PubMed] [Google Scholar]
- [28].Lau J, Ioannidis JPA, Balk EM, et al. Diagnosing acute cardiac ischemia in the emergency department: a systematic review of the accuracy and clinical effect of current technologies. Ann Emerg Med 2001;37:453–60. [DOI] [PubMed] [Google Scholar]
- [29].Higgins JPT, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol 2005;58:882–93. [DOI] [PubMed] [Google Scholar]
- [31].Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med 2008;359:366–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Punnoose EA, Atwal S, Liu W, et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res 2012;18:2391–401. [DOI] [PubMed] [Google Scholar]
- [33].Ran R, Li L, Wang M, et al. Determination of EGFR mutations in single cells microdissected from enriched lung tumor cells in peripheral blood. Anal Bioanal Chem 2013;405:7377–82. [DOI] [PubMed] [Google Scholar]
- [34].Breitenbuecher F, Hoffarth S, Worm K, et al. Development of a highly sensitive and specific method for detection of circulating tumor cells harboring somatic mutations in non-small-cell lung cancer patients. PLoS ONE 2014;9:e85350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Marchetti A, Del Grammastro M, Felicioni L, et al. Assessment of EGFR mutations in circulating tumor cell preparations from NSCLC patients by next generation sequencing: toward a real-time liquid biopsy for treatment. PLoS ONE 2014;9:e103883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Earhart CM, Hughes CE, Gaster RS, et al. Isolation and mutational analysis of circulating tumor cells from lung cancer patients with magnetic sifters and biochips. Lab Chip 2014;14:78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sundaresan TK, Sequist LV, Heymach JV, et al. Detection of T790M, the acquired resistance EGFR mutation, by tumor biopsy versus noninvasive blood-based analyses. Clin Cancer Res 2016;22:1103–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yin J, Yin HL, Wang Y, et al. Magnetic isolation of circulating tumor cells and its application in EGFR-TKI therapy of lung cancer. Chin J Clin Lab Sci 2015;59:23. [Google Scholar]
- [39].Rolfo C, Castiglia M, Hong D, et al. Liquid biopsies in lung cancer: the new ambrosia of researchers. Biochim Biophys Acta 2014;1846:539–46. [DOI] [PubMed] [Google Scholar]
- [40].Ilie M, Long E, Butori C, et al. ALK-gene rearrangement: a comparative analysis on circulating tumour cells and tumour tissue from patients with lung adenocarcinoma. Ann Oncol 2012;23:2907–13. [DOI] [PubMed] [Google Scholar]
- [41].Mayo C, Bertran-Alamillo J, Molina-Vila MÁ, et al. Pharmacogenetics of EGFR in lung cancer: perspectives and clinical applications. Pharmacogenomics 2012;13:789–802. [DOI] [PubMed] [Google Scholar]
- [42].Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2005;3:225–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005;352:786–92. [DOI] [PubMed] [Google Scholar]
- [44].Jiang T, Zhou C. Clinical activity of the mutant-selective EGFR inhibitor AZD9291 in patients with EGFR inhibitor-resistant non-small cell lung cancer. Transl Lung Cancer Res 2014;3:370–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yu N, Zhou J, Cui F, et al. Circulating tumor cells in lung cancer: detection methods and clinical applications. Lung 2015;193:157–71. [DOI] [PubMed] [Google Scholar]
- [46].Masuda H, Zhang D, Bartholomeusz C, et al. Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res Treat 2012;136:331–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Uramoto H, Shimokawa H, Nagata Y, et al. EGFR-activating mutations are not present in breast tumors of Japanese patients. Anticancer Res 2010;30:4219–22. [PubMed] [Google Scholar]
- [48].Tilch E, Seidens T, Cocciardi S, et al. Mutations in EGFR, BRAF and RAS are rare in triple-negative and basal-like breast cancers from Caucasian women. Breast Cancer Res Treat 2014;143:385–92. [DOI] [PubMed] [Google Scholar]
- [49].Fusi A, Metcalf R, Krebs M, et al. Clinical utility of circulating tumour cell detection in non-small-cell lung cancer. Curr Treat Options Oncol 2013;14:610–22. [DOI] [PubMed] [Google Scholar]
- [50].Qiu M, Wang J, Xu Y, et al. Circulating tumor DNA is effective for the detection of EGFR mutation in non-small cell lung cancer: a meta-analysis. Cancer Epidemiol Biomarkers Prev 2015;24:206–12. [DOI] [PubMed] [Google Scholar]
- [51].Smith B, Selby P, Southgate J, et al. Detection of melanoma cells in peripheral blood by means of reverse transcriptase and polymerase chain reaction. Lancet 1991;338:1227–9. [DOI] [PubMed] [Google Scholar]
- [52].Marzese DM, Hirose H, Hoon DSB. Diagnostic and prognostic value of circulating tumor-related DNA in cancer patients. Expert Rev Mol Diagn 2013;13:827–44. [DOI] [PubMed] [Google Scholar]
- [53].Diaz LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Higgins J, Altman DG. Assessing risk of bias in included studies. Cochrane Handbook for Systematic Reviews of Interventions: Cochrane Book Series 2008;187–241. [Google Scholar]
- [55].Moher D, Dulberg CS, Wells GA. Statistical power, sample size, and their reporting in randomized controlled trials. Jama 1994;272:122–4. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


