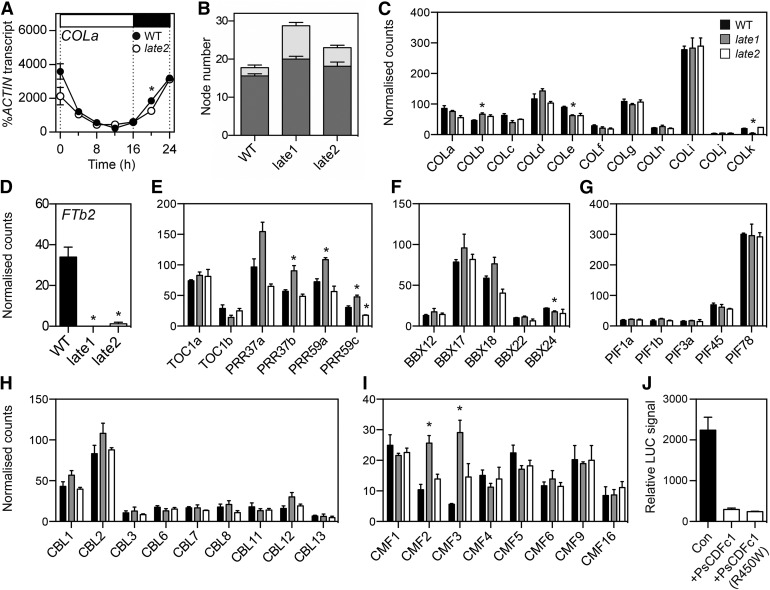
Figure 7.
Effects of LATE2 on FTb2 Expression Are Not Mediated by Expression Changes in Key Transcription Factor Families and May Result from Direct Transcriptional Repression.
(A) Expression of COLa in wild-type NGB5839 (WT) and late2-1D mutant plants under a 16-h LD photoperiod. Transcript levels were determined in the second uppermost fully expanded leaf of 3-week-old plants grown at 20°C. Mean values ± se are shown for n = 2 biological replicates, each consisting of pooled material from two plants. Day and night periods are indicated by white and black bars, respectively, above the graph.
(B) Flowering phenotypes of late1-2 and late2-1D mutants grown from sowing under continuous white light. Values represent mean ± se for n = 8 replicates. Dark gray, node of flower initiation; light gray, node of growth termination.
(C) to (I) Transcript levels of FTb2 (D) and genes in the COL (C), PRR (E), BBX (F), PIF (G), CBL (H), and CMF (I) families in expanded leaf tissue from wild-type, late1-2, and late2-1D mutants grown from sowing under continuous white light. Values represent mean ± se for n = 3 biological replicates, each consisting of pooled material from two plants. Asterisks indicate values significantly different from the wild type: P ≤ 0.05 for (A) and P ≤ 0.025 for (C) to (I).
(J) Luciferase activity driven from the M. truncatula FTb1 promoter:LUC construct coinfiltrated with Pro35S:GUS (control, denoted as “Con”), Pro35S:PsCDFc1 (wild type), or Pro 35S:PsCDFc1 (mutant R450W) and transiently expressed in N. benthamiana leaves. Data represents mean ± se for n = 8, 3, and 10 independently infiltrated plants, for control, PsCDFc1 wild-type, and PsCDFc1 mutant respectively.