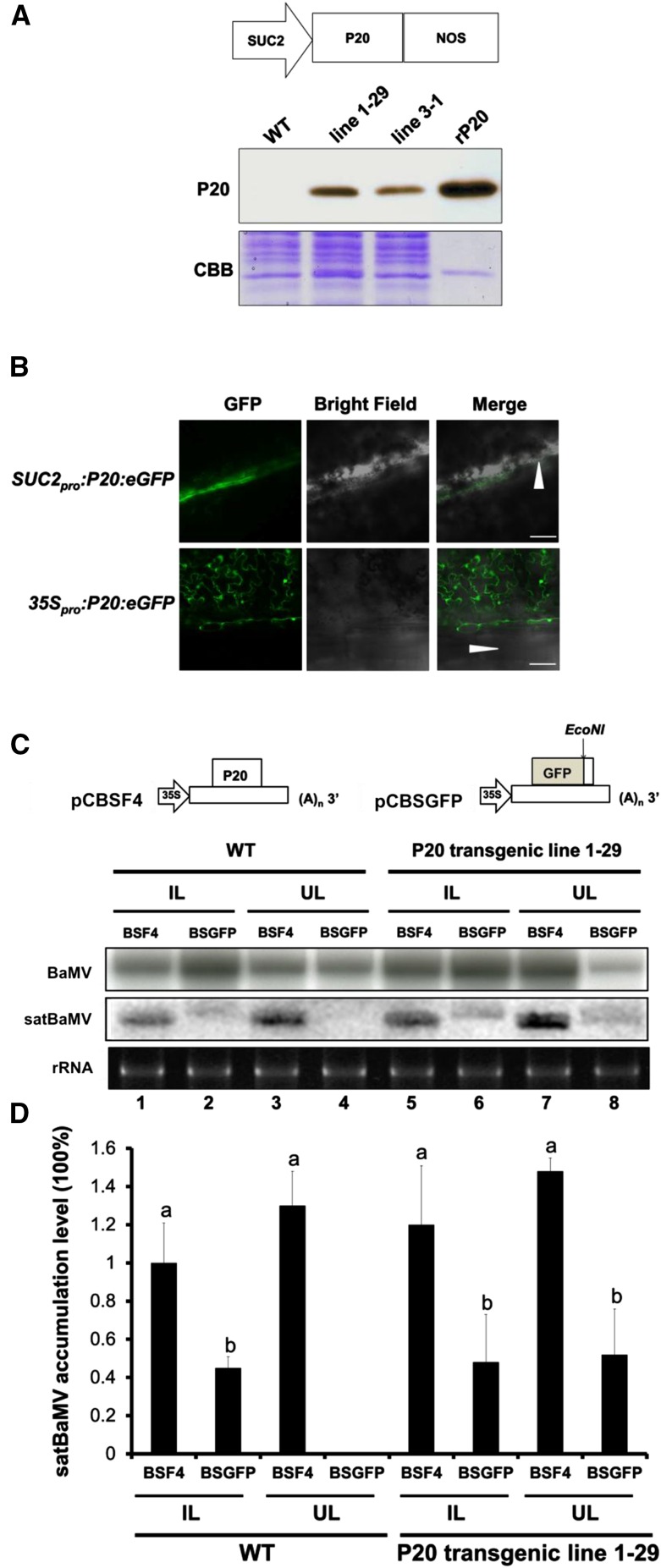
Figure 1.
Trans-Complementation of the Systemic Movement of P20-Defective satBaMV BSGFP in P20 Transgenic N. benthamiana.
(A) Physical map of a SUC2 promoter-driven P20 expression plasmid and immunoblot analysis of P20 accumulation in leaves of SUC2pro:P20 transgenic N. benthamiana (lines 1-29 and 3-1). rP20, recombinant P20 protein purified from Escherichia coli. CBB, Coomassie Brilliant Blue staining.
(B) P20-eGFP localization under SUC2 promoter- and 35S promoter-driven expression in wild-type N. benthamiana leaves at 3 DPI. Arrowheads represent the leaf midrib.
(C) Schematic maps of satBaMV infectious clones and RNA accumulation of wild-type satBaMV (BSF4) and P20-defective satBaMV (BSGFP) in wild-type and P20 transgenic N. benthamiana (line 1-29). Leaves of wild-type and P20-transgenic N. benthamiana were coinoculated with pCB (Lin et al., 2004) and pCBSF4 (Lin et al., 2004) or pCBSGFP. Inoculated leaves (IL) were harvested at 10 DPI and uninoculated upper leaves (UL) at 20 DPI for RNA gel blot analysis of BaMV and satBaMV RNA accumulation. BaMV and satBaMV accumulation was detected using 32P-labeled RNA probes specific for the BaMV 3′ end and satBaMV 3′-untranslated region, respectively.
(D) Quantitative analysis of satBaMV accumulation in wild-type and P20-transgenic line 1-29 from four independent biological samples, each involving four plants. Values are normalized against BSF4 satBaMV in inoculated leaves of wild-type plants. Data are mean ± sd from four experiments and were analyzed by Student’s t test. Different letters indicate significant difference (P < 0.05).