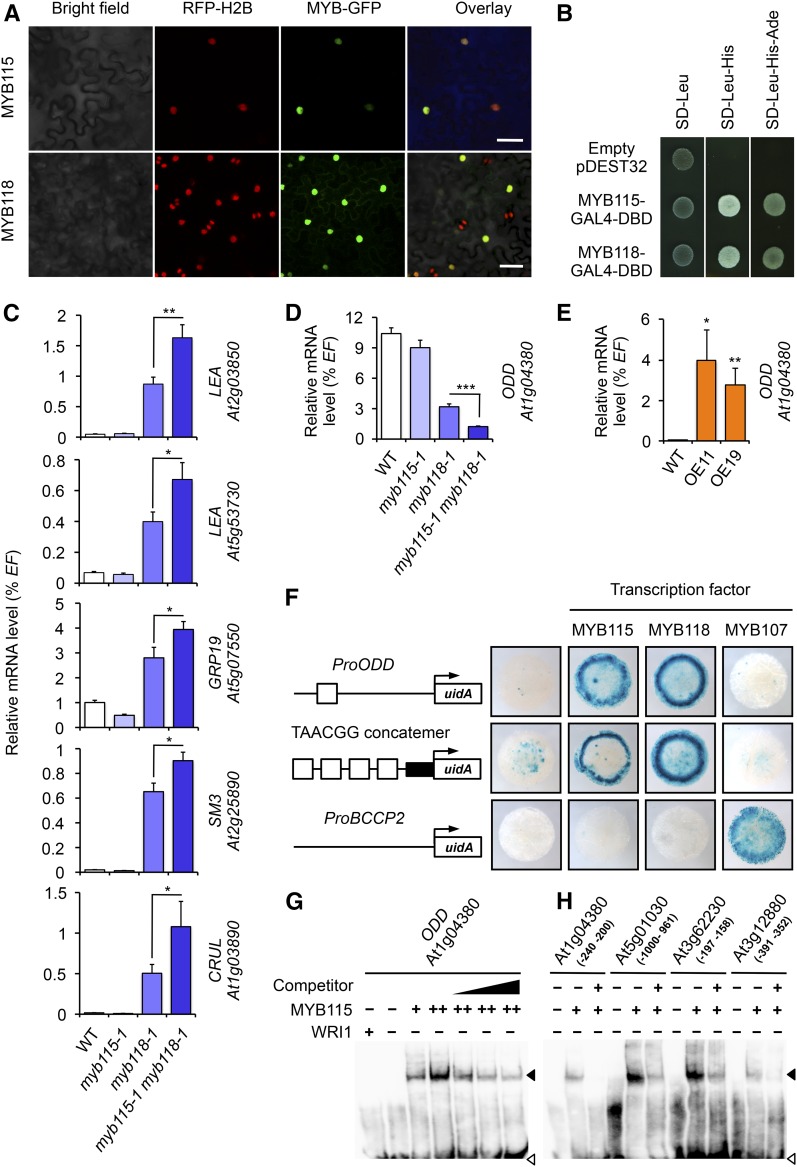
Figure 2.
Functional Characterization of MYB118 and MYB115.
(A) Confocal micrographs showing localization of MYB115:GFP and MYB118:GFP fusion proteins in transgenic N. benthamiana plants expressing RFP:H2B (Martin et al., 2009). Plants were coinfiltrated with the Pro35Sdual:MYB:GFP construct and a vector allowing the expression of the p19 protein of tomato bushy stunt virus (TBSV) that prevents the onset of posttranscriptional gene silencing (Shamloul et al., 2014). GFP was observed 4 d after infiltration. Bar = 50 µm.
(B) Transcriptional activity of MYB115 and MYB118. MYB115 and MYB118 coding sequences were cloned in frame with the GAL4 DNA binding domain (DBD). The fusion constructs were introduced into reporter yeast containing the HIS3 and ADE2 reporter genes, before being plated on appropriate media to maintain the expression of the vectors (SD-Leu) and to test the activation of the HIS3 (SD-Leu-His) or HIS3 and ADE2 reporter genes (SD-Leu-His-Ade). Data presented are representative from the results obtained for eight independent colonies. SD, synthetic dropout medium.
(C) and (D) RT-qPCR analysis of transcript abundance for negatively (C) and positively (D) regulated targets of MYB118 in cDNA prepared from wild-type (Col-0) and mutant seeds harvested 10 DAA. Values are the means and se of 12 replicates performed on cDNA dilutions obtained from three independent mRNA extractions. Asterisks indicate significant difference from the wild type according to t test at ***P < 0.001, ** P < 0.01, and * P < 0.05, respectively.
(E) RT-qPCR analysis of transcript abundance in cDNA prepared from rosette leaves of Pro35Sdual:MYB115 lines (OE11 and OE19). Values are the means and se of nine replicates performed on cDNA dilutions obtained from three independent mRNA extractions. Asterisks indicate significant difference from the wild type according to t test at **P < 0.01 and *P < 0.05, respectively.
(F) Transactivation assay in leaves of N. benthamiana. Schematic representations of the reporter constructs used are presented. Open boxes indicate TAACGG elements, and the closed box represents the 35S cauliflower mosaic virus minimal promoter. Pro:uidA reporter constructs alone or in combination with a vector allowing the expression of MYB115, MYB118, or MYB107 (negative control) were coinfiltrated in young leaves of N. benthamiana with a vector allowing the expression of the p19 protein. Leaf discs were assayed for GUS activity 3 d after infiltration. Tissues were incubated 17 h in a buffer containing 2 mM each of potassium ferrocyanide and potassium ferricyanide. Representative discs (diameter = 0.8 cm) are presented. TAACGG concatemer, promoter sequence made of a concatemer of TAACGG elements separated by 10 nucleotides (Barthole et al., 2014).
(G) Binding of MYB115 to the proximal upstream region of ODD. EMSA of a probe covering a region from −240 to −200 bp upstream from the ATG codon of ODD with increasing amounts of MYB115 (+ = 0.5 µg; ++ = 1.5 µg). WRI1 was used as a negative control. Competition of MYB115 binding was performed in the presence of 75-, 100-, and 200-fold amounts of the unlabeled ProODD (−240 to −200 bp) fragment. Position of free probe (open arrowhead) and the shifted bands (closed arrowhead) are indicated.
(H) Binding of MYB115 to the proximal upstream regions of targets of MYB118, namely, At5g01030, At3g62230, and At3g12880. The promoter sequence of the ODD gene (At1g04380) was used as a positive control. For each gene considered, the promoter region covered by the probe is indicated between parentheses.