TIC55, a Rieske-type oxygenase previously described as a component of the chloroplast protein import machinery, also functions as a chlorophyll catabolite hydroxylase.
Abstract
Chlorophyll degradation is the most obvious hallmark of leaf senescence. Phyllobilins, linear tetrapyrroles that are derived from opening of the chlorin macrocycle by the Rieske-type oxygenase PHEOPHORBIDE a OXYGENASE (PAO), are the end products of chlorophyll degradation. Phyllobilins carry defined modifications at several peripheral positions within the tetrapyrrole backbone. While most of these modifications are species-specific, hydroxylation at the C32 position is commonly found in all species analyzed to date. We demonstrate that this hydroxylation occurs in senescent chloroplasts of Arabidopsis thaliana. Using bell pepper (Capsicum annuum) chromoplasts, we establish that phyllobilin hydroxylation is catalyzed by a membrane-bound, molecular oxygen-dependent, and ferredoxin-dependent activity. As these features resemble the requirements of PAO, we considered membrane-bound Rieske-type oxygenases as potential candidates. Analysis of mutants of the two Arabidopsis Rieske-type oxygenases (besides PAO) uncovered that phyllobilin hydroxylation depends on TRANSLOCON AT THE INNER CHLOROPLAST ENVELOPE55 (TIC55). Our work demonstrates a catalytic activity for TIC55, which in the past has been considered as a redox sensor of protein import into plastids. Given the wide evolutionary distribution of both PAO and TIC55, we consider that chlorophyll degradation likely coevolved with land plants.
INTRODUCTION
Chlorophyll is the main pigment of the photosynthetic apparatus of plants and is responsible for the absorption of the energy of sunlight. In the process of photosynthesis, this energy is converted to chemical energy used for carbon fixation. However, during leaf senescence, when the photosynthetic machinery is dismantled in order to retrieve nutrients, in particular nitrogen, to sink organs such as seeds (Hörtensteiner and Feller, 2002), the photodynamic properties of chlorophyll become potentially cell toxic. As a consequence, the pigment is detoxified to uncolored and photodynamically inactive products that are termed phyllobilins (Kräutler, 2014). Chlorophyll degradation to phyllobilins is catalyzed by a multistep process, named the “PAO/phyllobilin” pathway (Hörtensteiner and Kräutler, 2011; Kräutler and Hörtensteiner, 2014), to acknowledge PAO (PHEOPHORBIDE a OXYGENASE), a Rieske-type oxygenase that is the key component of this pathway (Pruzinská et al., 2003). PAO, in concert with red chlorophyll catabolite reductase (RCCR), catalyzes the ring-opening reaction of pheophorbide a, the phytol- and Mg-free intermediate of the early part of chlorophyll breakdown, to a primary fluorescent chlorophyll catabolite (pFCC), the primary (fluorescent) phyllobilin that is the precursor of all subsequently formed phyllobilins. The direct product of the PAO reaction, red chlorophyll catabolite, does not accumulate but is immediately reduced at the C15/C16-double bond by RCCR in an intriguing stereospecific manner (Wüthrich et al., 2000; Pruzinská et al., 2007) (for atom and pyrrole numbering of phyllobilins, see the structure of pFCC in Figure 1B). Thus, depending on the source of the enzyme, two C16-stereoisomers, pFCC or epi-pFCC, may occur (Mühlecker et al., 1997, 2000). For example, the RCCR of Arabidopsis thaliana (At-RCCR) produces pFCC, while in bell pepper (Capsicum annuum) or tomato (Solanum lycopersicum), epi-pFCC is formed (Hörtensteiner et al., 2000). It was shown in vitro that the stereospecificity of Arabidopsis RCCR is defined by a single amino acid residue, Phe-219, which when mutated to Val changes the specificity (Pruzinská et al., 2007). Likewise, complementation of the accelerated cell death2 (acd2) mutant of Arabidopsis that is deficient in RCCR with an RCCR version “X,” where six amino acids of the wild-type sequence including Phe-219 were replaced by the respective residues of the tomato RCCR (acd2-2+At-RCCR-X), caused the exclusive accumulation during senescence of C16-isomerized phyllobilins (Pruzinská et al., 2007).
Figure 1.
pFCC and Hydroxy-pFCC Accumulate in Gerontoplasts of Arabidopsis.
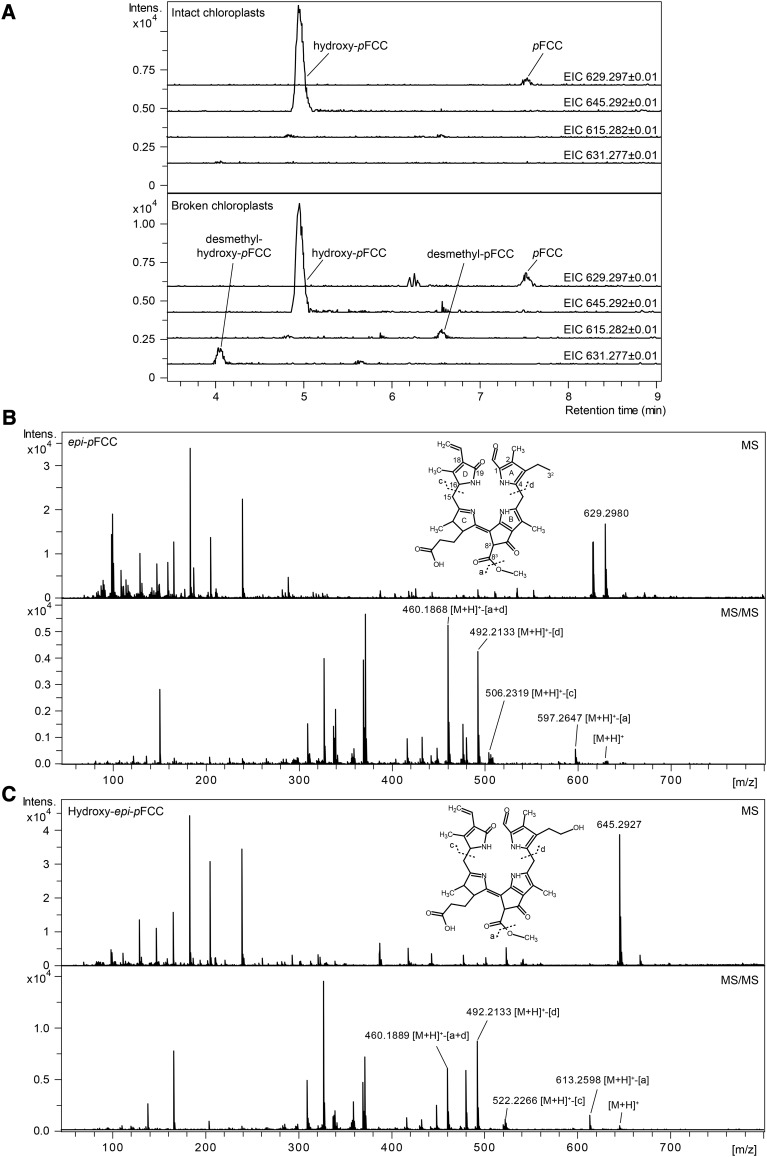
(A) Extracted ion chromatograms (EICs) of intact and broken gerontoplasts incubated with MES16. In intact gerontoplasts, pFCC (EIC 629.2977 ± 0.01) and hydroxy-pFCC (EIC 645.2929 ± 0.01) are not demethylated by the added MES16. However, in broken gerontoplasts, MES16 can access both as substrate, leading to the formation of demethylated products (EIC 615.2827 and EIC 631.2777, respectively). Thus, pFCC hydroxylation occurs inside the chloroplast.
(B) and (C) MS (top panel) and MS/MS spectra (bottom panel) of epi-pFCC (EIC 629.2980 ± 0.01; [B]) and hydroxy-epi-pFCC (EIC 645.2929 ± 0.01; [C]). Constitutional formulae and MS/MS fragmentation sites are shown. [M+H]+ indicates the precursor ion.
The end products of chlorophyll degradation are categorized into two types (Kräutler, 2014): (C1-) formyl-(C19-) oxobilins, also termed nonfluorescent chlorophyll catabolites (NCCs), are the ultimate products of PAO activity, while the activity of a cytochrome P450 monooxygenase, as shown for Arabidopsis (Christ et al., 2013), gives rise to (C1,19-) dioxobilins, also named dioxobilin-type nonfluorescent chlorophyll catabolites (DNCCs). NCCs and DNCCs are found in the vacuoles of senescent cells (Matile et al., 1988; Christ et al., 2012) and have been shown to be derived from the respective fluorescent precursor phyllobilins (fluorescent chlorophyll catabolites [FCCs] and dioxobilin-type fluorescent chlorophyll catabolites) as the result of nonenzymatic isomerization inside the acidic vacuolar sap (Oberhuber et al., 2003; Christ et al., 2013). Since the first identification of an NCC in senescent leaves of barley (Hordeum vulgare) in 1991 (Kräutler et al., 1991), phyllobilins were found in senescent leaves from more than 20 different angiosperm species (Kräutler, 2016) but are also formed during fruit ripening (Müller et al., 2007; Moser et al., 2009). Although the principal linear tetrapyrrole backbone of all phyllobilins is identical, they exhibit rather great structural diversity among the investigated species. Thus, for example, 90% of the phyllobilins that accumulate in senescent Arabidopsis leaves are DNCCs (Christ et al., 2013), while Cercidiphyllum japonicum exclusively produces NCCs (Oberhuber et al., 2003). Furthermore, some species, like Arabidopsis, accumulate an array of up to eight different phyllobilins simultaneously (Christ et al., 2016), while others, like C. japonicum, produce only two (Oberhuber et al., 2003). The molecular basis for these differences is enzyme activities that modify pFCC in a species-specific manner at different peripheral positions of the tetrapyrrole backbone structure. Best investigated in this respect is Arabidopsis where, besides CYP89A9, the already mentioned cytochrome P450 monooxygenase that is responsible for DNCC formation (Christ et al., 2013), methylesterase 16 (MES16) has been identified (Christ et al., 2012). This enzyme demethylates the C82 carboxymethyl ester present in chlorophyll with high efficiency. Again, other species, such as C. japonicum, exclusively produce phyllobilins that carry the intact carboxymethyl group, indicating that they lack a MES16 ortholog. Interestingly, these modifying reactions are localized in the cytosol (MES16) (Christ et al., 2012) and at the endoplasmic reticulum (CYP89A9) (Christ et al., 2013), while all enzymes identified to date that are required for the conversion of chlorophyll to pFCC are located within the plastid (Sakuraba et al., 2012). This led to the concept that reactions that commonly occur during chlorophyll breakdown take place in senescing chloroplasts, while species-specific modifications occur outside. Consequently, pFCC is believed to be exported from plastids, but the nature of pFCC transport at the plastid envelope is unknown (Christ and Hörtensteiner, 2014). An intriguing common modification of phyllobilins found in all species that have been analyzed so far is the specific hydroxylation at the C32 ethyl side chain (Christ and Hörtensteiner, 2014; Kräutler, 2014). This indicates that, distinct from other species-specific modifications, phyllobilin hydroxylation may be a reaction commonly occurring in angiosperms and that the activity may be localized in the plastid.
The aim of this work was to identify the molecular nature of the hydroxylating activity. We could show that, indeed, senescent chloroplasts (gerontoplasts) contained both pFCC and hydroxylated pFCC (hydroxy-pFCC), indicating the respective activity to be a chloroplast protein. Using bell pepper chromoplasts as starting material, we successfully established an in vitro enzyme assay that allowed us to biochemically characterize the properties of the enzyme in question. Based on these analyses and on 18O2 labeling and CO inhibition studies, we could narrow down the number of likely candidates to two Rieske-type oxygenases: TRANSLOCON AT THE INNER CHLOROPLAST ENVELOPE55 (TIC55) and PROTOCHLOROPHYLLIDE-DEPENDENT TRANSLOCON AT THE INNER CHLOROPLAST ENVELOPE52 (PTC52), which have both been implicated in chloroplast protein import. Analysis of respective T-DNA insertion mutants verified that tic55 mutants are unable to produce hydroxylated phyllobilins, while in a ptc52 mutant, hydroxylation was unaffected. Thus, TIC55 is responsible for phyllobilin hydroxylation during chlorophyll breakdown. TIC55 orthologs are widely distributed in higher plants but are phylogenetically distinct to PAO orthologs. This indicates that phyllobilin hydroxylation likely appeared with the evolution of land plants.
RESULTS
Hydroxy-pFCC Is Present in Gerontoplasts of Arabidopsis
The steps of chlorophyll to pFCC conversion have been shown to take place in gerontoplasts (Sakuraba et al., 2012). In agreement with this, fluorescent HPLC fractions corresponding to pFCC and to epi-pFCC have been identified in extracts of gerontoplasts of oilseed rape (Brassica napus) and barley (Schellenberg et al., 1993; Ginsburg et al., 1994; Moser and Matile, 1997). However, these early analyses indicated additional, more polar fluorescent fractions to be present in plastids. To investigate which, if any, chlorophyll catabolites are found in Arabidopsis plastids, isolated gerontoplasts were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Indeed, besides pFCC that produced a pseudomolecular ion of [M+H]+ = C35H41N4O7 (m/z 629.2977) in positive ionization mode, we identified a second, more polar fraction with a mass of m/z 645.2929 ([M+H]+ = C35H41N4O8), i.e., containing one additional oxygen atom (Figure 1A). In tandem MS experiments (Figures 1B and 1C), both ions fragmented in an identical manner that earlier had been shown to be characteristic for pFCC fragmentation (Mühlecker et al., 1997, 2000). Thus, pyrrole ring A (cleavage “d” shown in Figures 1B and 1C) and/or the C83 methoxy group (cleavage “a” shown in Figures 1B and 1C; loss as methanol) were lost with high efficiency, while pyrrole ring D was stable in both ions. To confirm the identity of these compounds as FCCs that are distinct from the respective NCCs, we performed isomerization experiments to experimentally convert an FCC to its respective NCC isomer (Oberhuber et al., 2003). For this, we used epi-pFCC that we produced from pheophorbide a in a well established in vitro system using bell pepper chromoplasts (Christ et al., 2012). On reverse-phase HPLC, epi-pFCC has a longer retention time than pFCC (Figure 2C) (Rodoni et al., 1997), but the fragmentation behavior in tandem MS experiments with a stable ring D was identical to that of pFCC (Supplemental Figure 1A). When incubating at pH 5, epi-pFCC isomerized to a more polar compound (Supplemental Figure 1B), which when analyzed by tandem MS fragmented as a typical NCC (Christ et al., 2016), i.e., with high probability of ring D loss (Supplemental Figure 1A), confirming the identity of the catabolites isolated from senescent Arabidopsis chloroplasts as FCCs, specifically as pFCC and hydroxy-pFCC, which solely differ by the attachment in the latter of an additional oxygen atom at ring A, likely at the C32 ethyl side chain.
Figure 2.
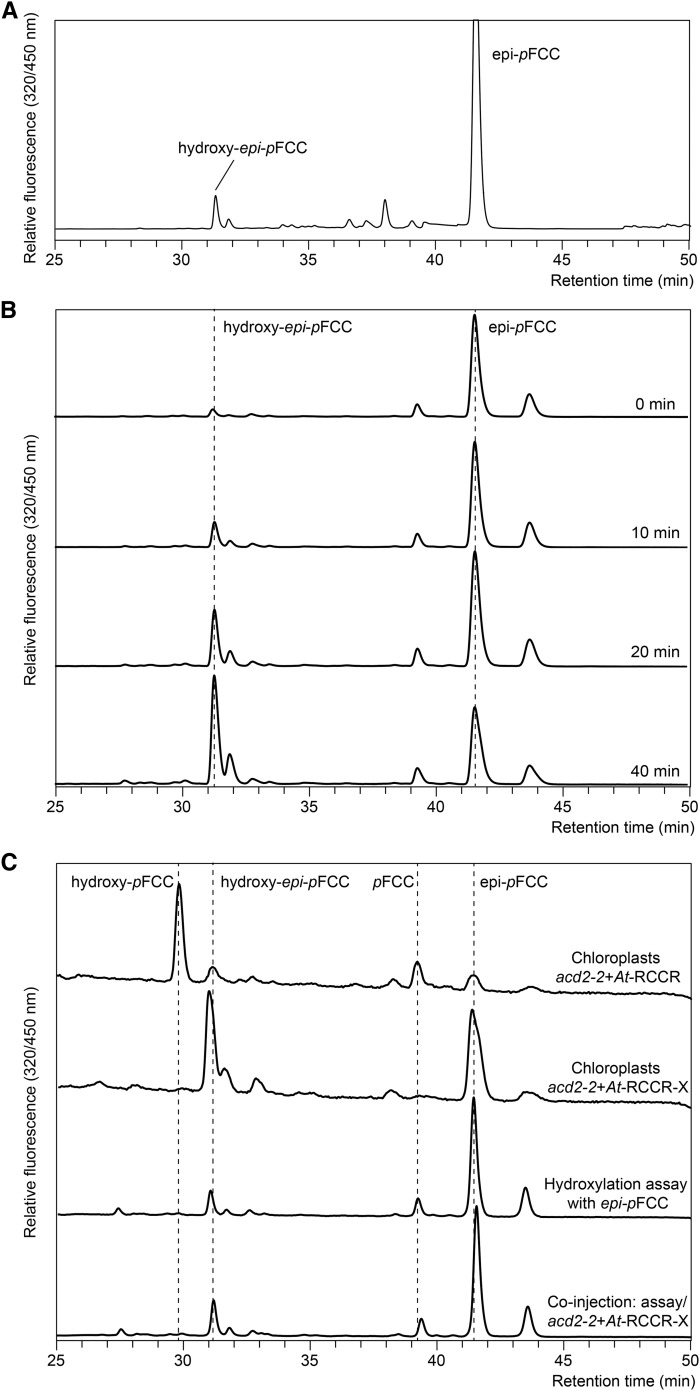
In Vitro Formation of Hydroxy-epi-pFCC from epi-pFCC in Bell Pepper Chromoplast Extracts.
(A) In vitro assay for the production of epi-pFCC from pheophorbide a using a bell pepper chromoplast extract. Besides epi-pFCC, the HPLC chromatogram shows a small, polar peak that was identified as hydroxy-epi-pFCC.
(B) Time-dependent synthesis of hydroxy-epi-pFCC from epi-pFCC in an assay that is identical to the one used in (A) for the production of epi-pFCC from pheophorbide a.
(C) HPLC-based confirmation of the product of the in vitro hydroxylation assay as hydroxy-epi-pFCC. Chromatograms from top: gerontoplast extracts from acd2-2+At-RCCR (Pruzinská et al., 2007) that produces pFCC and hydroxy-pFCC; gerontoplast extracts from acd2-2+At-RCCR-X (Pruzinská et al., 2007) that produces epi-pFCC and hydroxy-epi-pFCC; in vitro assay after 40 min of incubation; coinjection of acd2-2+At-RCCR-X and the assay, yielding a single polar peak that corresponds to hydroxy-epi-pFCC.
To verify that hydroxy-pFCC was present, and thus also likely formed, inside the intact plastids and to exclude the possibility that the identified FCCs were derived from extraplastidial contaminations of our chloroplast preparations, we took advantage of the fact that MES16 demethylates FCCs (Christ et al., 2012). Thus, we incubated plastid preparations before and after mechanical rupture with MES16 and analyzed the formation of the respective demethylated forms of pFCC and hydroxy-pFCC by LC-MS/MS (Figure 1A). Only after chloroplast rupture were pseudomolecular ions ([M+H]+) corresponding to desmethyl-pFCC (m/z 615.282; C34H39N4O7) and desmethyl-hydroxy-pFCC (m/z 631.277; C34H39N4O8) produced, indicating that, indeed, hydroxy-pFCC formation from pFCC takes place within intact senescent plastids.
pFCC Hydroxylation Localizes in Plastid Membranes and Requires Molecular Oxygen and Ferredoxin
The above-mentioned in vitro assay that uses bell pepper chromoplasts as the source of PAO and RCCR for the production of epi-pFCC from pheophorbide a yielded, as a by-product, a second, more polar, fluorescing fraction (Figure 2A) that exhibited a typical FCC absorption spectrum (Supplemental Figure 2A), which we reckoned could be hydroxy-epi-pFCC, indicating that the chromoplast extracts used for epi-pFCC synthesis may also contain the activities required for catabolite hydroxylation. Indeed, in an identical assay, but using epi-pFCC instead of pheophorbide a as substrate, the polar FCC fraction increased in a time-dependent manner (Figure 2B). To confirm identity of this compound as hydroxy-epi-pFCC, we compared its retention time in HPLC with the retention times of the FCCs accumulating in gerontoplasts of Col-0 plants and of acd-22+At-RCCR-X plants (Pruzinská et al., 2007) that were complemented with the epi-pFCC-producing “X” version of Arabidopsis RCCR (see Introduction). Coinjection experiments indeed confirmed that the product of in vitro hydroxylation of epi-pFCC is identical to the polar FCC fraction from acd2-2+At-RCCR-X chloroplasts (Figure 2C). In addition, the product of in vitro hydroxylation was analyzed by LC-MS/MS (Supplemental Figures 2B and 2C), demonstrating it to be hydroxy-epi-pFCC with an identical mass and fragmentation pattern as shown for hydroxy-pFCC (Figure 1B).
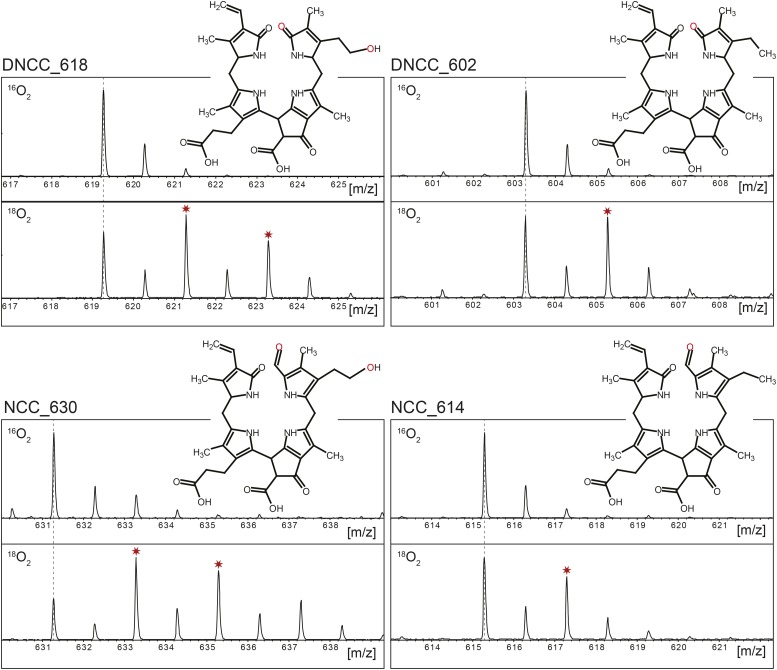
The C32 hydroxylation reaction introduces one oxygen atom into the respective phyllobilins. To analyze whether this oxygen was derived from molecular oxygen, we dark-incubated Arabidopsis wild-type leaves in the presence of 18O2 and analyzed phyllobilin labeling by LC-MS. Several phyllobilins known to be C32 hydroxylated, such as DNCC_618 and NCC_630 (Christ et al., 2016), carried up to two labeled oxygen atoms, while only one 18O-label was found in nonhydroxylated phyllobilins, such as DNCC_602 and NCC_614 (Figure 3). In all cases the labeled oxygen atoms were exclusively bound to pyrrole ring A. In the case of NCCs, one label was derived from PAO activity (Hörtensteiner et al., 1998), while CYP89A9 activity, ultimately leading to DNCCs, replaced the PAO-derived label by another labeled oxygen (Christ et al., 2013). This explained the presence of one 18O-label in nonhydroxylated phyllobilins, implying that the second label that was present in hydroxylated ones was derived by the activity of the hydroxylase, which introduced the oxygen atom at C32 from molecular oxygen.
Figure 3.
18O2-Labeling of Phyllobilins during Col-0 Leaf Senescence.
MS spectra of selected DNCCs (A) and NCCs (B) produced in the presence of ambient air (16O2; top spectra) or in an 18O2 atmosphere (bottom spectra). Note that in the presence of 18O2, C32-hydroxylated phyllobilins (e.g., DNCC_618 and NCC_630) carry two labeled oxygens, while nonhydroxylated phyllobilins (e.g., DNCC_602 and NCC_614) only carry one. This confirms that the C32 hydroxyl group oxygen is derived from molecular oxygen.
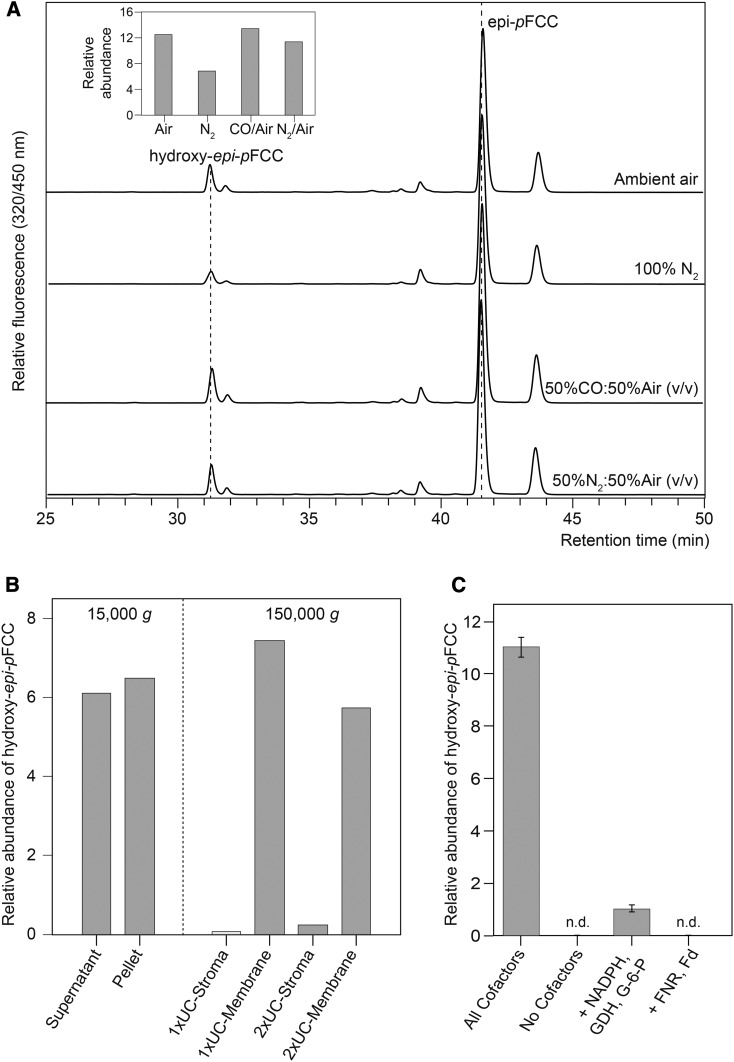
These data indicated that the hydroxylating activity could be a cytochrome P450 monooxygenase. Although most of these enzymes reside in the endoplasmic reticulum, the Arabidopsis genome encodes several P450 monooxygenases that likely are directed to the chloroplast (Schuler et al., 2006). P450 monooxygenases are highly sensitive to CO (Schuler, 1996), which we used in a mixture with 50% (v/v) ambient air to test for inhibition in the hydroxylation assay. However, CO did not inhibit hydroxy-epi-pFCC formation compared with an atmosphere containing a 1:1 (v/v) mixture of ambient air and nitrogen gas, while incubation in 100% nitrogen atmosphere largely inhibited the activity (Figure 4A). Thus, the responsible enzyme was unlikely a P450 monooxygenase.
Figure 4.
Biochemical Characterization of epi-pFCC Hydroxylation.
(A) In vitro hydroxylation assays with epi-pFCC as substrate were performed in different atmospheric conditions as indicated. Note that the absence of oxygen significantly inhibits hydroxy epi-pFCC formation (inset), thus confirming the molecular oxygen incorporation shown in Figure 3. Note also that CO treatment does not affect hydroxy epi-pFCC formation (inset), likely excluding the respective enzyme to be a cytochrome P450 monooxygenase. Inset: relative abundance of formed hydroxy epi-pFCC. Ratios of peak areas of hydroxy epi-pFCC and epi-pFCC are shown.
(B) Fractionation of the epi-pFCC hydroxylating activity in bell pepper extracts. Fractionation at low centrifugation speed (15,000g) into a soluble (Supernatant) and membrane (Pellet) fraction was unable to clearly separate the activity; however, subsequent twofold ultracentrifugation (UC) (150,000g) of the supernatant fraction uncovered the activity to reside in chromoplast membranes (1xUC- and 2xUC-Membrane), while no activity was found in respective supernatants (1xUC- and 2xUC-Stroma).
(C) epi-pFCC hydroxylation depends on ferredoxin. Cofactor requirement was analyzed in assays using the 2xUC-Membrane fraction of (B) and added factors as indicated. All cofactors are Fd, NADPH, G-6-P, and GDH. Note that in the absence of Fd little activity was retained, likely because of the presence of residual amounts of Fd in the 2xUC-Membrane fraction. Data are the mean of three replicates. Error bars indicate sd.
The bell pepper chromoplast-based in vitro assay for epi-pFCC production from pheophorbide a was shown to require both chromoplast membranes, as the source of PAO, and a soluble chromoplast fraction, as the source of RCCR. To further dissect the requirements for in vitro epi-pFCC hydroxylation, we fractionated total chromoplasts into soluble and membrane fractions. After two washing steps using ultracentrifugation, the activity remained in the membrane fraction, indicating that the responsible enzymes are tightly associated with chloroplast membranes (Figure 4B). Similarly, the in vitro epi-pFCC hydroxylation assay was analyzed for the requirement for the additional cofactors present in the original PAO/RCCR assay, i.e., ferredoxin (Fd) and a Fd-reducing system, consisting of NADPH, Fd-NADPH oxidoreductase (FNR), glucose-6-phosphate (G-6-P), and G-6-P dehydrogenase (GDH). Using 2× ultracentrifugation-washed membranes to reduce carryover of potential cofactors, respective assays for the production of hydroxy epi-pFCC demonstrated the requirement of both Fd and the Fd-regenerating system (Figure 4C). Some activity was obtained without the addition of Fd, which likely was caused by some residual Fd remaining attached to the chromoplast membranes during the washing procedure.
pFCC Hydroxylation Is Catalyzed by TIC55, a Rieske-Type Monooxygenase
The properties for the in vitro epi-pFCC hydroxylating activity determined above were similar to the requirements of PAO, a Rieske-type monooxygenase (Schellenberg et al., 1993; Hörtensteiner et al., 1995, 1998); i.e., activity was attached to plastid membranes, was dependent on Fd, and was not inhibited by CO, and the incorporated oxygen atom was derived from molecular oxygen. The Arabidopsis genome encodes five Rieske-type oxygenases (Gray et al., 2004), all of which have been shown to localize to chloroplasts (Rathinasabapathi et al., 1997; Tanaka et al., 1998; Bartsch et al., 2008). In addition, three of them, PAO (At3g44880), TIC55 (At2g24820), and PTC52 (At4g25650), possess predicted C-terminally located transmembrane domains (Figure 5A) that likely anchor these proteins to plastid membranes, while the other two Rieske-type oxygenases in Arabidopsis, chlorophyll a oxygenase (At1g44446) and choline monooxygenase (CMO)-like (At4g29890) are predicted to be soluble proteins (Tusnády and Simon, 2001). Both TIC55 and PTC52 have been considered to be components of distinct protein import complexes of the chloroplast, i.e., the TIC and PTC complex, respectively; however the role of the PTC complex in protein import in true leaves has been questioned (Kim and Apel, 2004). Nevertheless, we considered both proteins as likely candidates for FCC hydroxylation. First, we reanalyzed their predicted localization in the envelope (Bartsch et al., 2008) by transiently expressing GFP fusion proteins in mesophyll protoplasts isolated from senescent Arabidopsis wild-type leaves (Figure 5B). Indeed, GFP signals for both TIC55 and PTC52 surrounded the chloroplasts, i.e., indicating envelope localization like the envelope positive control, TIC110, while PAO colocalized with chlorophyll autofluorescence, i.e., confirming thylakoid localization (Sakuraba et al., 2012).
Figure 5.
Protein Alignment and Analysis of Subcellular Localization of the Arabidopsis Rieske-Type Oxygenases TIC55, PTC52, and PAO.
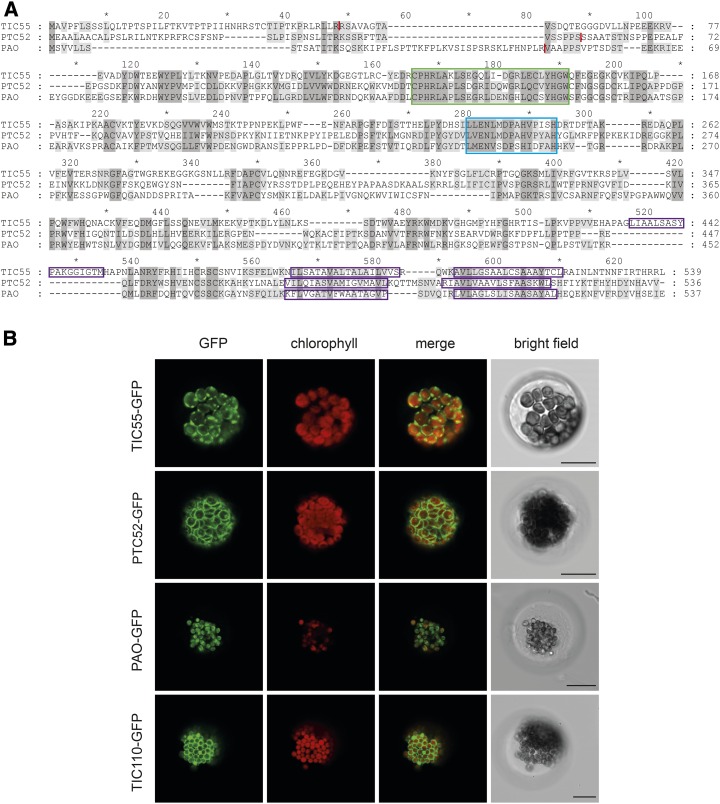
(A) Sequence alignment of Arabidopsis TIC55, PTC52, and PAO. Sequences were aligned using the program DIALIGN. Dark-gray shading highlights residues identical between all three proteins, and light-gray shading highlights residues identical between two proteins, with Blosum62 similarity groups enabled. The following colored domains are boxed: green, Rieske center; blue, mononuclear iron site; purple, predicted transmembrane domains. The predicted cleavage sites of chloroplast transit peptides are indicated with red lines.
(B) Localization of Rieske-type oxygenase-GFP fusion proteins in protoplasts isolated from senescent Arabidopsis wild-type leaves. GFP fluorescence (column 1) and chlorophyll autofluorescence (column 2) were examined by confocal laser scanning microscopy. Column 3, merge of GFP and chlorophyll fluorescence; column 4, bright-field image. Bars = 20 μm.
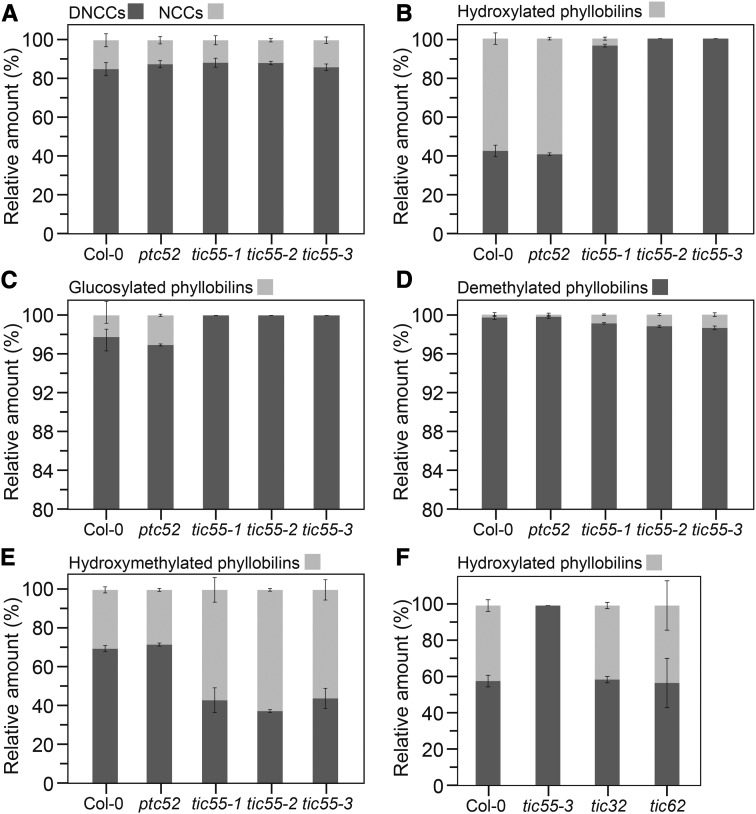
To investigate whether one (or both) of these candidates catalyzes FCC hydroxylation in vivo, we analyzed respective T-DNA insertion lines for both genes. As shown previously (Boij et al., 2009), none of the lines showed growth differences compared with the wild type. In addition and similar to mutants in phyllobilin-modifying enzymes (Christ et al., 2012, 2013), we did not detect a visible phenotype during senescence. However, LC-MS analysis of senescent leaf extracts uncovered major phyllobilin differences between the three analyzed tic55 alleles and ptc52, which looked like the wild type (Supplemental Figure 3). Although the relative abundance of DNCCs and NCCs was unaltered in all investigated lines (Figure 6A), two of the three tic55 mutants, tic55-2 and tic55-3, lacked hydroxylated phyllobilins (Figure 6B). The tic55-1 mutants had a largely reduced proportion of hydroxylated phyllobilins, implying some residual activity to be present in this allele. By contrast, ptc52 was comparable to Col-0 with ∼60% of all phyllobilins being hydroxylated. Furthermore, glucosylated phyllobilins were absent from tic55 mutants (Figure 6C), showing that phyllobilin glucosylation depends on prior C32 hydroxylation. These results demonstrated that phyllobilin hydroxylation is catalyzed by TIC55, but not by PTC52. To our surprise, tic55 mutants accumulated a smaller proportion of O84 demethylated phyllobilins (Figure 6D), indicating that MES16, catalyzing demethylation in Arabidopsis (Christ et al., 2012), might preferentially accept hydroxylated catabolites as substrate. Furthermore, the proportion of hydroxymethylated phyllobilins was higher in tic55 mutants (Figure 6E), which was in agreement with the suggestion that C2 or C4 hydroxymethylation and C32 hydroxylation exclude each other (Süssenbacher et al., 2015).
Figure 6.
Phyllobilin Analysis of tic and ptc52 Mutants.
Abundance of phyllobilins was determined by LC-MS/MS analysis according to a published method (Christ et al., 2016) and is shown as relative values. Data are the mean of four biological replicates. Error bars indicate sd.
(A) to (E) Phyllobilin abundance in short-day-grown tic55 and ptc52 mutants after senescence induction for 5 d. Note the different scale used in (C) and (D) because of the low abundance of glucosylated and demethylated phyllobilins.
(F) Abundance of hydroxylated phyllobilins in a tic32 and tic62 mutant.
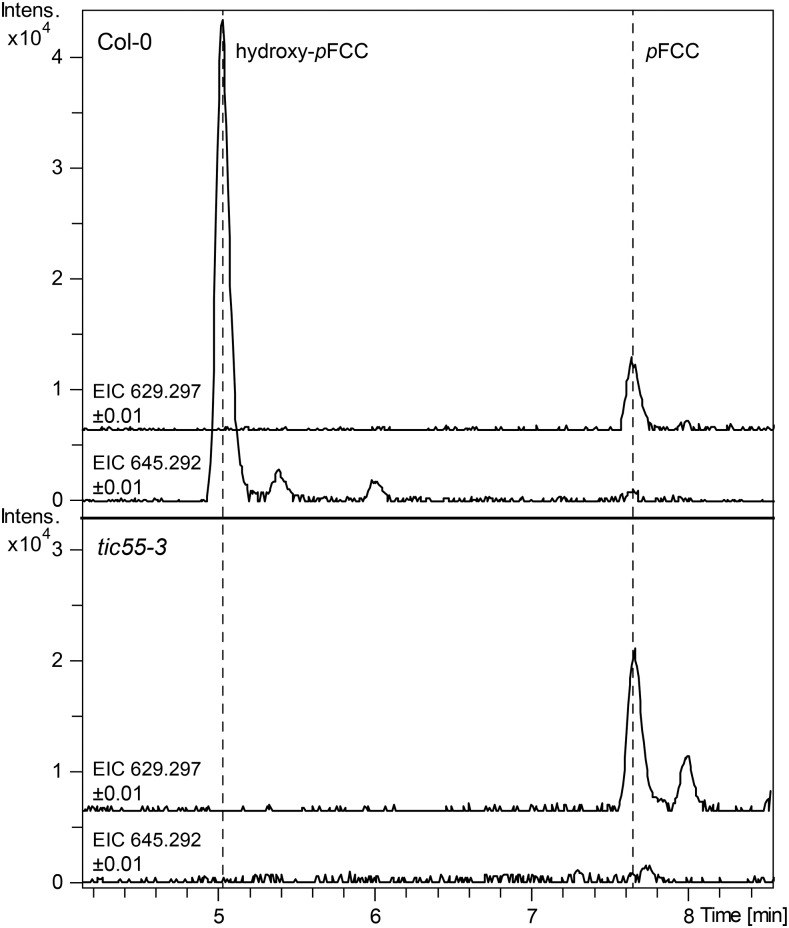
We additionally confirmed the absence of pFCC hydroxylation in tic55-3 by analyzing the FCC pattern of isolated plastids. As expected, hydroxy-pFCC was absent from gerontoplasts, which contained relatively more pFCC (Figure 7).
Figure 7.
Hydroxy-pFCC Is Absent from tic55-3 Gerontoplasts.
Extracted ion chromatograms (EICs) of pFCC (EIC 629.2977 ± 0.01) and hydroxy-pFCC (EIC 645.2929 ± 0.01) of gerontoplasts of the wild type (Col-0) and tic55-3 are shown. Note the absence of hydroxy-pFCC from the mutant.
TIC32 and TIC62 Are Not Involved in the Redox Cycle of TIC55
TIC55 had been considered to exhibit a redox regulatory role in protein import together with TIC32 and TIC62, two additional proposed components of the protein import machinery at the inner chloroplast envelope (Küchler et al., 2002; Hörmann et al., 2004). Both proteins were shown to possess NADPH binding domains and, in addition, TIC62 is able to interact with FNR (Bölter et al., 2015). This implies that one or both of these additional TIC components could be involved in delivering electrons to the Rieske and/or the mononuclear iron center of TIC55 and thus drive the redox cycle required for TIC55 activity. To test this possibility, we analyzed phyllobilin accumulation in tic32 and tic62 T-DNA insertion mutants during leaf senescence. As seen in Figure 6F, mutations in these genes did not affect phyllobilin hydroxylation, indicating that electron supply toward TIC55 is independent from TIC62 or TIC32.
DISCUSSION
Phyllobilin Hydroxylation by TIC55
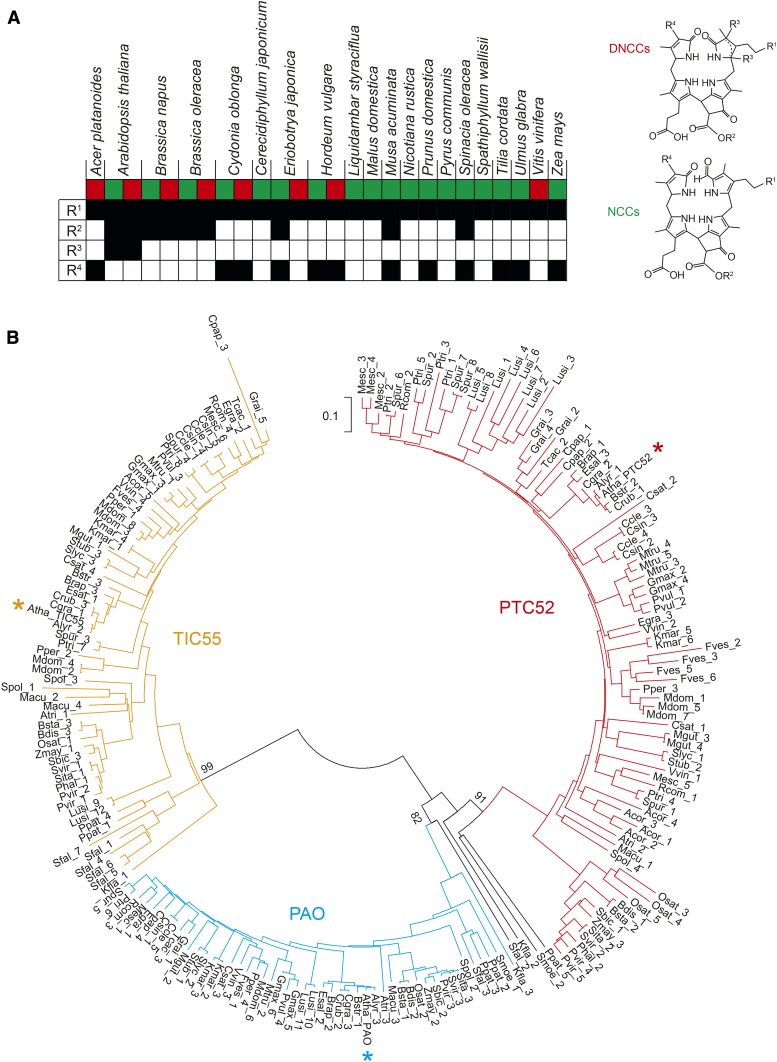
Most of the phyllobilins that have been identified to date as degradation products of chlorophyll in higher plants carry a hydroxyl group at the C32 position (Christ and Hörtensteiner, 2014; Kräutler, 2014, 2016; Scherl et al., 2016). Furthermore, hydroxylated phyllobilins have been found in each species analyzed so far (Figure 8A) and in many cases have been shown to represent the major phyllobilin fraction (Oberhuber et al., 2003; Christ et al., 2013). This implies that chlorophyll catabolite hydroxylation is a common and important reaction, which, together with the upstream reactions that lead to the formation of pFCC, forms the core part of the PAO/phyllobilin pathway of chlorophyll breakdown in higher plants. Here, we identified TIC55, an envelope-localized member of the Rieske-type oxygenases, as the phyllobilin hydroxylase in Arabidopsis. Our biochemical characterization and subsequent analysis of mutants defective in likely candidates unequivocally demonstrates that TIC55 is solely responsible for the formation of hydroxylated phyllobilins during leaf senescence-related chlorophyll breakdown. More specifically, based on the localization of TIC55 in the chloroplast and the fact that other phyllobilin-modifying activities are located outside plastids (Christ et al., 2012, 2013), pFCC is the likely sole in vivo substrate for hydroxylation by TIC55.
Figure 8.
Phyllobilin Distribution and Phylogenetic Analysis of Rieske-Type Oxygenases in Plants.
(A) Phyllobilin hydroxylation (R1) is a common modification, which is found in DNCCs (red square) and NCCs (green square) of all 20 species analyzed so far for structures of phyllobilins. By contrast, other modifications (R2-R4; see DNCC and NCC structures for the position of these modifications) occur in a species-specific manner.
(B) Phylogenetic analysis of Arabidopsis TIC55 (yellow asterisk), PAO (blue asterisk), and PTC52 (red asterisk) homologs in plants. Note that except for a few (black lines), most of the 204 protein sequences compared in this analysis cluster in one of three distinct clades. Also note that for each species at least one Rieske-type oxygenase protein is located within each clade. The tree was generated with the neighbor-joining method using MEGA7 (Kumar et al., 2016). Important nodes separating the three clades are labeled with bootstrap values (% of 1000 replicates). Acor, Aquilegia coerulea; Alyr, Arabidopsis lyrata; Atha, Arabidopsis thaliana; Atri, Amborella trichopoda; Bdis, Brachypodium distachon; Brap, Brassica rapa; Bsta, Brachypodium stacei; Bstr, Boechera stricta; Ccle, Citrus clementina; Cgra, Capsella grandiflora; Cpap, Carica papaya; Crub, Capsella rubella; Csat, Cucumis sativus; Csin, Citrus sinensis; Egra, Eucalyptus grandis; Esal, Eutrema salsugineum; Fves, Fragaria vesca; Gmax, Glycine max; Grai, Gossypium raimondii; Kfla, Klebsormidium flaccidum; Kmar, Kalanchoe marnieriana; Lusi, Linum usitatissimum; Macu, Musa acuminata; Mdom, Malus domstica; Mesc, Manihot esculenta; Mgut, Mimulus guttatus; Mtru, Medicago truncatula; Osat, Oryza sativa; Phal, Panicum hallii; Ppat, Physcomitrella patens; Pper, Prunus persica; Ptri, Populus trichocarpa; Pvir, Panicum virgatum; Pvul, Phaseolus vulgaris; Rcom, Ricinus communis; Sbic, Sorghum bicolor; Sfal, Sphagnum fallax; Sita, Setaria italica; Slyc, Solanum lycopersicum; Some, Selaginella moellendorffii; Spol, Spirodela polyrhiza; Spur, Salix purpurea; Stub, Solanum tuberosum; Svir, Setaria viridis; Tcac, Theobroma cacao; Vvin, Vitis vinifera; Zmay, Zea mays.
This finding is rather surprising because TIC55 was originally identified as a potential component of the protein import machinery at the chloroplast inner envelope (Caliebe et al., 1997). Specifically, it was proposed that TIC55 together with TIC62 and TIC32 functions as redox sensor that links protein import to the redox state of the chloroplast (Balsera et al., 2010). As such, TIC55, TIC62, and TIC32 would play a regulatory role rather than being core components of the inner envelope import machinery itself, the exact composition of which is still under debate (Bölter et al., 2015). The rationale behind this proposed function is the presence in TIC55 of redox-active cysteine residues at the C terminus that have been shown to be regulated by the thioredoxin system (Bartsch et al., 2008). Furthermore, the other two components of the so-called redox regulon (Stengel et al., 2007), i.e., TIC62 and TIC32, possess NADPH binding sites and could exert their regulatory role depending on changes of the NADP+/NADPH ratio, a well known readout of the redox state of the chloroplast. Indeed, TIC62 was shown to shuttle between thylakoids and the inner envelope in a NADP+/NADPH ratio-dependent manner (Stengel et al., 2008), which together with its additional ability to bind FNR (Benz et al., 2009) could allow electron fluxes at these membranes to be highly dynamically regulated (Bölter et al., 2015). Metabolic NADP+/NADPH ratios directly influenced import efficiency of a subset of preproteins in pea (Pisum sativum; Stengel et al., 2009). In addition, inhibition of protein import by ionomycin and ophiobolin A, two drugs that disrupt calcium signaling, has been attributed to inhibition of TIC32, which possesses a calmodulin binding site (Chigri et al., 2006). However, mutant analysis of redox regulon components did not show clear phenotypes, implying that under standard conditions, this redox control may be not essential for plant viability and/or chloroplast protein import (Benz et al., 2009; Boij et al., 2009; Balsera et al., 2010).
Here, we show that TIC55 exhibits a defined role as phyllobilin hydroxylase; however, ferredoxin-derived electrons that are required to drive the redox cycle of this Rieske-type oxygenase (Ferraro et al., 2005) unlikely involve TIC62 and/or TIC62-bound FNR because respective mutants did not exhibit phyllobilin hydroxylation deficiency.
Rieske-Type Oxygenases Play a Dominant Role in Chlorophyll Metabolism
As stated above, the role of TIC55 in protein import has not been firmly established. The Rieske-type iron-sulfur cluster and the mononuclear iron site present in this protein were considered as parts of a hypothetical electron transport chain at the inner envelope with potential final transfer of electrons to oxygen, thus integrating protein import to the redox regulatory network of chloroplasts (Bölter et al., 2015). However, potential substrates for TIC55 that, as a genuine Rieske-type oxygenase (Ferraro et al., 2005), catalyze activation and incorporation of molecular oxygen have neither been considered nor identified. Our 18O2 labeling experiments of senescent leaves combined with in vitro activity assays demonstrate that TIC55 is a monooxygenase that incorporates one oxygen atom derived from molecular oxygen into pFCC.
The Arabidopsis genome encodes five Rieske-type oxygenases (Gray et al., 2002). With the identification of TIC55 as a phyllobilin hydroxylase reported here, four out of these five have been attributed to chlorophyll metabolism. Two of them are involved in chlorophyll biosynthesis, i.e., CAO that catalyzes the oxidation of chlorophyll(ide) a to chlorophyll(ide) b (Tanaka et al., 1998), and PTC52 that has been proposed to carry out the same oxidation, but using protochlorophyllide a as substrate. However, an in vivo role of protochlorophyllide b has been questioned (Scheumann et al., 1999; Armstrong et al., 2000). The other two Rieske-type oxygenases are involved in chlorophyll degradation, i.e., PAO, which is responsible for porphyrin ring opening of pheophorbide a (Pruzinská et al., 2005), and TIC55. The fifth Rieske-type oxygenase, CMO-like, has high sequence similarity to CMO of spinach (Spinacia oleracea) that catalyzes the first step of glycine betaine biosynthesis (Rathinasabapathi et al., 1997). However, Arabidopsis does not produce glycine betaine and Arabidopsis CMO-like was unable to promote glycine betaine formation (Hibino et al., 2002).
Phylogenetic analyses had revealed that for all five Rieske-type oxygenases highly homologous proteins are present in higher plants (Gray et al., 2004). Reevaluation of the homologies of PAO, PTC52, and TIC55 incorporating sequences from Klebsormidium flaccidum (Hori et al., 2014) and from the Phytozome database (https://phytozome.jgi.doe.gov), which currently includes 52 sequenced Viridiplantae species, showed that these three Rieske-type oxygenase cluster into three distinct clades (Figure 8B). TIC55 and PAO orthologs are present in all land plants, including Selaginella moellendorffii, a member of the Lycopodiopsida, the oldest lineage of vascular plants, and the bryophyte Physcomitrella patens. The wide distribution of all three homologs within land plant species, but apparent absence from chlorophyte green algae for which genomes are available, such as Chlamydomonas reinhardtii, Micromonas pusilla, Volvox carteri, and Ostreococcus lucimarinus, suggests that TIC55 in higher plants most likely originated from a recent gene duplication and subsequent neofunctionalization to play a potential role in the adaptation to a terrestrial growth habit and/or high light adaptation. Interestingly, the genome of the early charophyte species K. flaccidum, which is considered to be a close ancestor of land plants (Hori et al., 2014), also encodes TIC55- and PAO-like proteins that exhibit rather high sequence similarities to the respective proteins from land plants. This is in support of the idea that chlorophyll degradation coevolved with the colonization of land by plants.
Spatial Distribution of Chlorophyll Catabolic Enzymes within the Plastid
The common reactions of the PAO/phyllobilin pathway leading to pFCC localize to the chloroplast. This has been substantiated by numerous investigations using different approaches, among them GFP fusion protein analysis, subcellular fragmentation studies, and enzyme activity measurements on isolated chloroplast fractions (for recent reviews on the biochemistry of chlorophyll breakdown, see Hörtensteiner, 2013a, 2013b). However, the subchloroplast location of respective reactions remained largely unclear until recently, in particular because of the inconsistency with regard to the presence or absence of transmembrane domains in these proteins. Thus, for example, PAO is an integral membrane protein with two predicted transmembrane domains (Figure 5A) (Sakuraba et al., 2012), while RCCR, with which it physically interacts during catalysis (Rodoni et al., 1997; Pruzinská et al., 2007), is a soluble protein (Wüthrich et al., 2000). Recently, based on protein interaction studies, we proposed the model that all chloroplast-located chlorophyll catabolic enzymes form a highly dynamic multiprotein complex at the thylakoid membrane with interaction with light harvesting complex II proteins (Sakuraba et al., 2012).
The identification of the envelope-bound TIC55 as the phyllobilin hydroxylase is intriguing in this respect (see Figure 9 for a topographical model of the PAO/phyllobilin pathway integrating TIC55). Nevertheless, one can speculate that there is no requirement of including TIC55 in the above-mentioned thylakoid membrane-bound degradative complex because the proposed rationale behind this complex, i.e., metabolic channeling of degradation intermediates to prevent potential phototoxicity (Sakuraba et al., 2012), is reached with the formation of pFCC. A pFCC derivative was recently shown to be able to efficiently produce singlet oxygen in vitro when excited with UV light (Jockusch et al., 2014). Nevertheless, when compared with pheophorbide, which absorbs a much wider spectral range and is known as being highly cell phototoxic (Xodo et al., 2012), the in vivo capacity of pFCC to produce reactive oxygen species and, thus, to act as a phototoxin is most likely rather limited. Envelope localization of TIC55 implies the requirement for pFCC movement from the thylakoid to the envelope, but it remains unclear whether this is achieved by simple diffusion or is protein-guided. Instead of being important for intermediate detoxification, like the upstream reactions, the localization of TIC55 at the envelope rather implies a link to catabolite export from the plastid. The molecular nature of the chloroplast exporter(s) for chlorophyll catabolites is unknown to date, but it has been speculated to be driven by an active transport mechanism (Matile et al., 1992). The former link of TIC55 with protein import at the inner envelope (Küchler et al., 2002) implies the intriguing possibility for phyllobilin export through the same protein complex; however, this is highly speculative and export through the TIC/TOC system has never been described. Whether TIC55 may directly interact with potential catabolite exporters remains unclear because phyllobilin export does not absolutely require previous reaction by TIC55, as demonstrated by the presence of a substantial proportion of nonhydroxylated phyllobilins in senescent wild-type leaves (Figure 6B) (Christ et al., 2016)
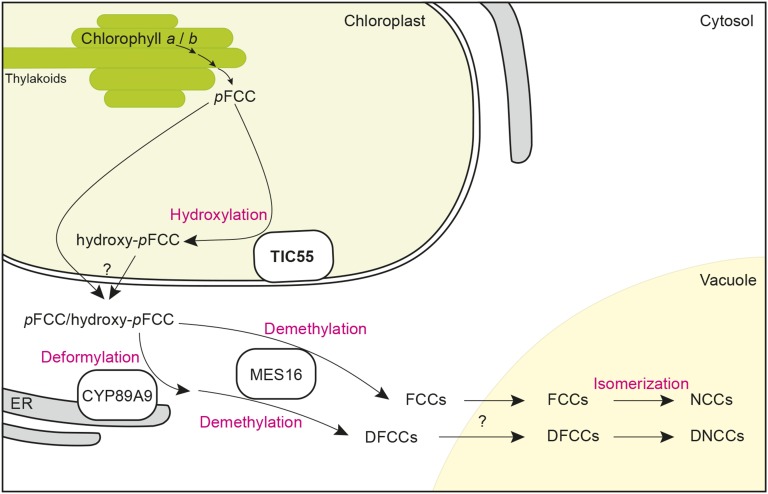
Figure 9.
Topographical Model of the Chlorophyll Breakdown Pathway Integrating the Findings of This Work.
The model shows the subcellular localization of pFCC-modifying activities, i.e., CYP89A9 (deformylation), MES16 (demethylation), and TIC55 (hydroxylation). Note that all the reactions involved in converting chlorophyll to pFCC, including PAO the name-giving key enzyme of the PAO/phyllobilin pathway, are not shown but have been considered to form a highly dynamic complex at the thylakoid membrane to prevent release into the stroma of potentially phototoxic breakdown intermediates upstream of pFCC (Sakuraba et al., 2012). Note also that due to the fact that nonhydroxylated phyllobilin occur in wild-type Arabidopsis, a fraction of pFCC likely leaves the chloroplast without hydroxylation. As indicated by the question marks, the molecular identity of phyllobilin transporters at the chloroplast envelope and the tonoplast is unknown. DFCC, dioxobilin-type fluorescent chlorophyll catabolites; ER, endoplasmic reticulum; pFCC, primary fluorescent chlorophyll catabolite.
What Is the Biological Role of Phyllobilin Modification?
None of the Arabidopsis mutants that are deficient in one of the known phyllobilin-modifying reactions, i.e., CYP89A9, MES16, or TIC55, show an obvious visible phenotype or are compromised in chlorophyll breakdown during senescence (Christ et al., 2012, 2013). Furthermore, deformylation (through CYP89A9 in Arabidopsis) and demethylation (through MES16 in Arabidopsis), but also C18-dihydroxylation and C32-OH group conjugation, are species-specific reactions (Hörtensteiner and Kräutler, 2011; Kräutler, 2014). Thus, seemingly these are nonessential reactions in chlorophyll degradation. Nevertheless, the fact that all three, i.e., TIC55, CYP89A9, and MES16, are members of protein subfamilies with three, seven, and three members, respectively, in Arabidopsis (Gray et al., 2004; Bak et al., 2011; Christ et al., 2012), suggests that they have been specifically recruited during evolution to function in chlorophyll breakdown. This implies that phyllobilins could possibly play a role beyond being merely degradation products. In line with this, phyllobilins have been shown to be efficient antioxidants and may act as light filters or optical brighteners (Kräutler, 2016). To what extent such physicochemical properties may serve for a biological role and to what extent phyllobilin modification is important in this respect remain unclear so far. The identification of TIC55 in this work offers a tool for producing a cyp89a9 mes16 tic55 triple mutant in the future, for a more in-depth analysis about the role of phyllobilin modification during chlorophyll breakdown and to investigate whether absence of these modifications might have an impact on plant fitness, for example, during stress conditions.
METHODS
Plant Material and Senescence Induction
Arabidopsis thaliana ecotype Col-0 was used as the wild type. T-DNA insertion lines for the following genes were used: TIC55 (AT2G24820), PTC52 (AT4G25650), TIC32 (At4g23430), and TIC62 (At3g18890). They were from the following collections: SALK lines (Alonso et al., 2003): tic55-1, SALK_137948; tic55-2, SALK_086048C; and ptc52, SALK_006984; GABI-KAT lines (Rosso et al., 2003): tic32, GK_117H08; and tic62, GK_439H04; SAIL line (Sessions et al., 2002): tic55-3, SAIL_167_G08. These lines were obtained from the European Arabidopsis Center, Nottingham, UK. Homozygous plants were identified by PCR using T-DNA- and gene-specific primers as listed in Supplemental Table 1. Two Arabidopsis acd2-2 complementation lines that express RCCR with either stereospecificity (acd2-2+At-RCCR, producing pFCC; and acd2-2+At-RCCR-X, producing epi-pFCC) (Pruzinská et al., 2007) were used as reference for HPLC analysis.
For biochemical and physiological experiments, plants were either grown under short-day conditions (16 h dark/8 h light) or under long-day conditions (16 h light/8 h dark) under fluorescent light of 60 to 120 μmol photons m−2 s−1 at 22°C. Plants for seed production and DNA isolation were grown in a greenhouse with rates of 100 to 200 μmol photons m−2 s−1 at 22°C. Leaf senescence was inducted by incubating detached leaves on wet filter paper for up to 8 d in permanent darkness. For dark incubation under 18O2 atmosphere, leaves from short-day-grown Col-0 plants were excised and placed on wet filter paper in a glass desiccator. After evacuation with a vacuum pump, the desiccator was aerated with 18O2 (97.1%; Campro Scientific) and leaves senesced in the dark for 5 d. Red bell pepper (Capsicum annuum) fruits used for chromoplast isolation were bought from a local market.
Biocomputational Methods
Orthologs of TIC55, PAO, and PTC52 were identified using BLASTP searches (Altschul et al., 1997) at the Phytozome database of the Joint Genome Institute (https://phytozome.jgi.doe.gov). Klebsormidium flaccidum sequences were likewise obtained from the National Center for Biotechnology Information (http://ncbi.nlm.nih.gov). For phylogenetic analysis, a multiple sequence alignment was generated with the neighbor-joining method using MEGA7 (Kumar et al., 2016). Bootstrap analysis was performed with 1000 replicates.
GFP Fusion Protein Production and Confocal Microscopy
TIC55 (BX819909) and PTC52 (pda06002) full-length cDNAs were obtained from the Centre National de Ressources Génomiques Végétales, France, and the RIKEN Tsukuba Institute BioResource Center, Japan, respectively. TIC55 was amplified by PCR with KAPA HiFi HotStart ReadyMix (Kapa Biosystems) using the primers listed in Supplemental Table 1. The resulting PCR fragment was cut with XmaI and SpeI and ligated to XmaI/SpeI-digested pUC18-spGFP6 (Meyer et al., 2006) to create a fusion between TIC55 and GFP at the C terminus. Likewise, PTC52 was amplified by PCR using the primers listed in Supplemental Table 1 and cloned into pUC18-spGFP6 via BamHI. Correctness of cDNA sequences and orientation was verified by sequencing. As references for localization, GFP fusion proteins of TIC110 (chloroplast inner envelope membrane) and PAO (thylakoid membrane) (Sakuraba et al., 2012) were used. The constructs were used to transform Arabidopsis protoplasts isolated from 6-week-old plants grown in short-day conditions. Isolation of protoplasts was performed according to a published protocol (Christ et al., 2012, and references therein). GFP fluorescence was imaged with a confocal laser scanning microscope (TCS SP5; Leica Microsystems) at an excitation wavelength of 488 nm, and signals were detected between 495 and 530 nm. Chlorophyll autofluorescence was recorded at 643 to 730 nm.
Arabidopsis Chloroplast Isolation and MES16 Incubation
Four- to six-week-old plants grown in short-day conditions were used for chloroplast isolation in a two-step purification procedure. A published protocol (Schelbert et al., 2009) was adapted to optimize isolation of senescent protoplasts and chloroplasts. Leaves were detached and dark incubated for 4 d. After isolation, mesophyll protoplasts were lysed by the addition of breakage buffer (containing 300 mM sorbitol, 20 mM Tricine-KOH, pH 8.4, 5 mM EDTA, 5 mM EGTA, 10 mM NaHCO3, and 0.1% [w/v] BSA) and gerontoplasts purified on a discontinuous (85 and 40% [v/v]) gradient of Percoll containing the same salts as the breakage buffer. After washing the chloroplasts obtained from the 85%/40% interphase twice with HEPES-sorbitol buffer (330 mM sorbitol and 50 mM HEPES-KOH, pH 8), they were analyzed for colorless catabolites by HPLC or by LC-MS/MS (see below). For this, chloroplast fractions were supplemented with one volume of methanol (v/v), mixed by vortexing, and twice centrifuged (16,000g; 2 min). Alternatively, isolated chloroplasts were incubated for 10 min with MES16. For this, recombinant MES16 was produced as described (Christ et al., 2012). A twenty-fifth volume of MES16-containing crude protein extract (1 µg µL−1) was either added directly to intact chloroplasts or to chloroplasts that were broken up by freezing in liquid N2 prior to the addition of the enzyme. Reactions were stopped by the addition of one volume of methanol (v/v) and vortexing. After centrifugation twice (16,000g; 2 min), the formation of demethylated hydroxy-pFCC was analyzed by LC-MS/MS (see below).
epi-pFCC Preparation
In order to produce epi-pFCC as substrate for the hydroxylation assay, PAO and RCCR were extracted from red bell pepper chromoplasts according to a published protocol (Christ et al., 2012). The extracted enzymes were then used to convert pheophorbide a (Hörtensteiner et al., 1995) to epi-pFCC according to an established assay (Pruzinská et al., 2007). epi-pFCC was concentrated on C18-SepPak columns (Waters) and purified by HPLC (see below) for later use in hydroxylation assays.
Isolation of the Hydroxylating Activity from Bell Pepper Chromoplasts and Activity Determination
Total chromoplasts isolated from red bell pepper fruits were also used as source of the hydroxylating enzyme. Essentially the same protocol as for the extraction of PAO and RCCR (Christ et al., 2012) was used, but with some modifications. Briefly, red bell pepper exocarp was blended with a fruit juicer in chromoplast isolation buffer (1.33 mL g−1 tissue; Christ et al., 2012), filtered through Miracloth, and then centrifuged for 10 min at 12,000g. The pellet was carefully resuspended in chromoplast isolation buffer (0.33 mL g−1 tissue; Christ et al., 2012) and recentrifuged. The pellet was finally resuspended in 8 mL of 25 mM Tris-MES, pH 8.0 (25 μL g−1 tissue), and chromoplasts were broken by pressing them 10 times through a 0.6-mm syringe needle. The resulting total chromoplast protein fraction was either frozen in liquid nitrogen and stored at −80°C for later use or centrifuged (15,000g, 10 min) to separate supernatant and pellet. The supernatant was further fractionated by ultracentrifugation (UC) at 150,000g for 60 min into 1xUC-Membrane and 1xUC-Stroma. The obtained pellet was resuspended in the start volume of 25 mM Tris-MES, pH 8.0, and 10 times pushed through a needle as above. The ultracentrifugation step was repeated, yielding 2xUC-Membrane and 2xUC-Stroma fractions, respectively.
The standard hydroxylation assays (total volume 50 µL) consisted of 28 μL of respective chromoplast fractions, 15 µM of epi-pFCC, 10 µg of Fd (Sigma-Aldrich), and a Fd-reducing system consisting of 10 mM G-6-P, 2.5 mM NADPH, 50 milliunits GDH, and 5 milliunits of FNR (Sigma-Aldrich). After incubation at 25°C in darkness for up to 40 min, the reactions were terminated by adding methanol to a final concentration of 50% (v/v). After centrifugation for 2 min at 16,000g, samples were analyzed by HPLC or by LC-MS/MS as described below. For reactions under defined atmospheric conditions, assays were set up on ice in glass bulbs. After removal of the ambient air by vacuum suction, bulbs were flushed with the desired gas mixtures and assays performed as described above.
Colorless Chlorophyll Catabolites
Chlorophyll catabolites were either analyzed by HPLC or by LC-MS/MS.
HPLC
For HPLC analysis, plant material was ground in liquid nitrogen and colorless catabolites were extracted with 3 volumes (w/v) of 50 mM phosphate buffer pH 7/methanol (1:3, v/v). Plant extracts or hydroxylation assay reactions were analyzed on a reverse-phase HPLC system as described (Christ et al., 2012). The column was developed with a gradient (flow rate 1 mL min−1) of solvent A (25 mM potassium phosphate, pH 7.0), solvent B (methanol), and solvent C (H2O) as follows (%A/%B/%C [v/v/v]): from 80/20/0 to 40/60/0 in 35 min, to 0/60/40 in 12 min, to 0/100/0 in 2 min, back to 0/60/40 in 7 min, and finally back to 80/20/0 in 2 min. Peak detection was performed with sequential monitoring using a PDA-100 photodiode array detector (200 to 700 nm; ThermoFisher Scientific) and a RF2000 fluorescence detector (excitation at 320 nm, emission at 450 nm; Shimadzu). Chlorophyll catabolites were identified by their absorption (FCCs, NCCs, and DNCCs) and fluorescence (FCCs) properties. Relative amounts of hydroxy-epi-pFCC obtained in hydroxylation assays were determined by peak areas at 320 nm.
LC-MS/MS
For LC-MS/MS analysis, plant material was collected in 2-mL microfuge tubes containing 500 μL of 1.25- to 1.65-mm glass beads. The tissue was ground in liquid nitrogen using a MM300 Mixer Mill (Retsch) at 30 Hz for 5 min. Chlorophyll catabolites were extracted with 5 volumes (w/v) of ice-cold 80% methanol, 20% water, and 0.1% formic acid (v/v/v) containing 1 µg mL−1 ampicillin as internal standard, sonicated for 2 min, and twice centrifuged at 16,000g for 2 min. Total plant extracts, chloroplast extracts, or reactions of hydroxylation assays were analyzed by LC-MS/MS (Thermo Fisher Scientific UltiMate 3000 RSLC system, coupled to a Compact Q-TOF mass spectrometer with electron spray ionization [Bruker Daltonics]) as described (Christ et al., 2016). Two different LC programs were employed. In the short program (Christ et al., 2016), the column was developed with a gradient (flow rate of 0.3 mL min−1) of solvent B (acetonitrile with 0.1% [v/v] formic acid) in solvent A (water with 0.1% [v/v] formic acid) as follows (all v/v): 30% for 0.5 min, 30 to 70% in 7.5 min, 70 to 100% in 0.1 min, and 100% for 3.9 min. In the long program, identical solvents were used, but the column was developed as follows (all v/v): 5% B for 0.5 min, 30% B to 100% B in 11 min, and 100% B for 4 min. Phyllobilins were identified and quantified using an in-house-built spectrum library (Christ et al., 2016).
Accession Numbers
The following genes were used: TIC55 (AT2G24820), PTC52 (AT4G25650), TIC32 (At4g23430), and TIC62 (At3g18890). Sequence data from K. flaccidum can be found in GenBank/EMBL database (http://www.ncbi.nlm.nih.gov) under the following accession numbers: Kfla_1 (GAQ77973.1), Kfla_2 (GAQ78701.1), and Kfla_3 (GAQ92150.1). All other sequences can be found in the Phytozome database (https://phytozome.jgi.doe.gov) under the following accession numbers: Aquilegia coerulea: Acor_1, 22029521; Acor_2, 22030025; Acor_3, 22030648; Acor_4, 22035222; Acor_5, 22062530; Arabidopsis lyrata: Alyr_1, 16058352; Alyr_2, 16049304; Alyr_3, 16052333; Arabidopsis thaliana: Atha_PAO (At3g44880), 19664072; Atha_PTC52 (At4g25650), 19646489; Atha_TIC55, 19,643,456 (At2g24820); Amborella trichopoda: Atri_1, 31571943; Atri_2, 31573466; Atri_3, 31569219; Brachypodium distachon: Bdis_1, 32798942; Bdis_2, 32805471; Bdis_3, 32814680; Brassica rapa: Brap_1, 30640093; Brap_2, 30639463; Brap_3, 30615756; Brachypodium stacei: Bsta_1, 32873310; Bsta_2, 32877401; Bsta_3, 32863896; Boechera stricta: Bstr_1, 30657466; Bstr_2, 30657466; Bstr_3, 30677801; Citrus clementina: Ccle_1, 20794617; Ccle_2, 20794921; Ccle_3, 20814974; Ccle_4, 20817945; Ccle_5, 20814172; Capsella grandiflora: Cgra_1, 28896530; Cgra_2, 28912086; Cgra_3, 28914744; Carica papaya: Cpap_1, 16406577; Cpap_2, 16406579; Cpap_3, 16409268; Cpap_4, 16420643; Capsella rubella: Crub_1, 20894180; Crub_2, 20886114; Crub_3, 20902616; Cucumis sativus: Csat_1, 16963640; Csat_2, 16963642; Csat_3, 16968781; Csat_4, 16973514; Citrus sinensis: Csin_1, 18104396; Csin_2, 18110147; Csin_3, 18110062; Csin_4, 18109308; Csin_5, 18109304; Eucalyptus grandis: Egra_1, 32059933; Egra_2, 32036721; Egra_3, 32033828; Eutrema salsugineum: Esal_1, 20208666; Esal_2, 20196368; Esal_3, 20194144; Fragaria vesca: Fves_1, 27274244; Fves_2, 27249443; Fves_3, 27248027; Fves_4, 27263237; Fves_5, 27276459; Fves_6, 27275924; Glycine max: Gmax_1, 30490036; Gmax_2, 30523624; Gmax_3, 30553241; Gmax_4, 30540276; Gmax_5, 30528498; Gmax_6, 30548653; Gossypium raimondii: Grai_1, 26820373; Grai_2, 26773144; Grai_3, 26773258; Grai_4, 26773690; Grai_5, 26765361; Kalanchoe marnieriana: Kmar_1, 32592111; Kmar_2, 32568622; Kmar_3, 32585756; Kmar_4, 32565330; Kmar_5, 32587661; Kmar_6, 32537639; Linum usitatissimum: Lusi_1, 23170108; Lusi_10, 23152413; Lusi_11, 23179897; Lusi_12, 23153476; Lusi_2, 23178747; Lusi_3, 23178807; Lusi_4, 23178789; Lusi_5, 23178765; Lusi_6, 23150802; Lusi_7, 23150844; Lusi_8, 23150790; Lusi_9, 23156438; Musa acuminata: Macu_1, 32305345; Macu_2, 32298593; Macu_3, 32299412; Macu_4, 32301167; Malus domestica: Mdom_1, 22648216; Mdom_2, 22680745; Mdom_3, 22673860; Mdom_4, 22657421; Mdom_5, 22669270; Mdom_6, 22681531; Mdom_7, 22646192; Mdom_8, 22657424; Manihot esculenta: Mesc_1, 32328964; Mesc_2, 32352206; Mesc_3, 32352768; Mesc_4, 32351848; Mesc_5, 32351381; Mesc_6, 32343636; Mimulus guttatus: Mgut_1, 28939106; Mgut_2, 28929299; Mgut_3, 28927831; Mgut_4, 28928063; Medicago truncatula: Mtru_1, 31054512; Mtru_2, 31108864; Mtru_3, 31076177; Mtru_4, 31075891; Mtru_5, 31071508; Oryza sativa: Osat_1, 33136036; Osat_2, 33129257; Osat_3, 33130101; Osat_4, 33126987; Osat_5, 33128763; Panicum hallii: Phal_1, 32526147; Phal_2, 32510109; Physcomitrella patens: Ppat_1, 32958411; Ppat_2, 32971600; Ppat_3, 32943162; Ppat_4, 32923430; Ppat_5, 32934884; Prunus persica: Pper_1, 32111912; Pper_2, 32118911; Pper_3, 32087889; Pper_4, 32086408; Populus trichocarpa: Ptri_1, 27044612; Ptri_2, 27044125; Ptri_3, 26998722; Ptri_4, 26997353; Ptri_5, 26997432; Ptri_6, 26992599; Ptri_7, 27007363; Ptri_8, 27010289; Panicum virgatum: Pvir_1, 30249395; Pvir_2, 30232924; Pvir_3, 30210200; Pvir_4, 30208474; Pvir_5, 30224983; Phaseolus vulgaris: Pvul_1, 27170687; Pvul_2, 27168327; Pvul_3, 27147757; Pvul_4, 27151382; Ricinus communis: Rcom_1, 16802948; Rcom_2, 16802951; Rcom_3, 16803313; Rcom_4, 16811159; Sorghum bicolor: Sbic_1, 32753731; Sbic_2, 32754631; Sbic_3, 32744056; Sphagnum fallax: Sfal_1, 32628626; Sfal_2, 32612026; Sfal_3, 32611292; Sfal_4, 32631199; Sfal_5, 32631202; Sfal_6, 32631211; Sfal_7, 32615809; Setaria italica: Sita_1, 32697922; Sita_2, 32693755; Sita_3, 32689656; Solanum lycopersicum: Slyc_1, 27278224; Slyc_2, 27297008; Slyc_3, 27308810; Selaginella moellendorffii: Smoe_1, 15421380; Smoe_2, 15420984; Spirodela polyrhiza: Spol_1, 31503412; Spol_2, 31521506; Spol_3, 31512183; Spol_4, 31510559; Salix purpurea: Spur_1, 31446831; Spur_2, 31444937; Spur_3, 31408599; Spur_4, 31425200; Spur_5, 31434385; Spur_6, 31420119; Spur_7, 31420805; Spur_8, 31420645; Solanum tuberosum: Stub_1, 24409796; Stub_2, 24385728; Stub_3, 24377864; Setaria viridis: Svir_1, 32663617; Svir_2, 32653526; Svir_3, 32653709; Theobroma cacao: Tcac_1, 27426931; Tcac_2, 27427235; Tcac_3, 27426954; Vitis vinifera: Vvin_1, 17833437; Vvin_2, 17833438; Vvin_3, 17837646; Vvin_4, 17841070; Zea mays: Zmay_1, 30990042; Zmay_2, 31009998; Zmay_3, 31,016,834.
Supplemental Data
Supplemental Figure 1. Confirmation of epi-pFCC and pFCC Identity and epi-pFCC-to-epi-pNCC Isomerization.
Supplemental Figure 2. LC-MS Confirmation of Hydroxy-epi-pFCC.
Supplemental Figure 3. LC-MS Analysis of Phyllobilins from Col-0 and Rieske-Type Oxygenase Mutants.
Supplemental Table 1. List of Primers Used in This Study.
Supplemental File 1. Text File of the Alignment Corresponding to the Phylogenetic Analysis in Figure 8.
Supplementary Material
Acknowledgments
This work was supported by the Swiss National Science Foundation (Grant 31003A_149389) and CropLife (to S.H.).
AUTHOR CONTRIBUTIONS
S.H. designed research. M.H., B.C., and A.D. performed research. M.H., B.C., S.A., and S.H. analyzed data. M.H. and S.H. wrote the article.
Glossary
- RCCR
red chlorophyll catabolite reductase
- NCC
nonfluorescent chlorophyll catabolite
- DNCC
dioxobilin-type nonfluorescent chlorophyll catabolite
- FCC
fluorescent chlorophyll catabolite
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- FNR
Fd-NADPH oxidoreductase
- G-6-P
glucose-6-phosphate
- GDH
G-6-P dehydrogenase
- CMO
choline monooxygenase
Footnotes
Articles can be viewed without a subscription.
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong G.A., Apel K., Rüdiger W. (2000). Does a light-harvesting protochlorophyllide a/b-binding protein complex exist? Trends Plant Sci. 5: 40–44. [DOI] [PubMed] [Google Scholar]
- Bak S., Beisson F., Bishop G., Hamberger B., Höfer R., Paquette S., Werck-Reichhart D. (2011). Cytochrome p450. The Arabidopsis Book 9: e0144, doi/10.1199/tab.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsera M., Soll J., Buchanan B.B. (2010). Redox extends its regulatory reach to chloroplast protein import. Trends Plant Sci. 15: 515–521. [DOI] [PubMed] [Google Scholar]
- Bartsch S., Monnet J., Selbach K., Quigley F., Gray J., von Wettstein D., Reinbothe S., Reinbothe C. (2008). Three thioredoxin targets in the inner envelope membrane of chloroplasts function in protein import and chlorophyll metabolism. Proc. Natl. Acad. Sci. USA 105: 4933–4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz J.P., Stengel A., Lintala M., Lee Y.H., Weber A., Philippar K., Gügel I.L., Kaieda S., Ikegami T., Mulo P., Soll J., Bölter B. (2009). Arabidopsis Tic62 and ferredoxin-NADP(H) oxidoreductase form light-regulated complexes that are integrated into the chloroplast redox poise. Plant Cell 21: 3965–3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boij P., Patel R., Garcia C., Jarvis P., Aronsson H. (2009). In vivo studies on the roles of Tic55-related proteins in chloroplast protein import in Arabidopsis thaliana. Mol. Plant 2: 1397–1409. [DOI] [PubMed] [Google Scholar]
- Bölter B., Soll J., Schwenkert S. (2015). Redox meets protein trafficking. Biochim. Biophys. Acta 1847: 949–956. [DOI] [PubMed] [Google Scholar]
- Caliebe A., Grimm R., Kaiser G., Lübeck J., Soll J., Heins L. (1997). The chloroplastic protein import machinery contains a Rieske-type iron-sulfur cluster and a mononuclear iron-binding protein. EMBO J. 16: 7342–7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chigri F., Hörmann F., Stamp A., Stammers D.K., Bölter B., Soll J., Vothknecht U.C. (2006). Calcium regulation of chloroplast protein translocation is mediated by calmodulin binding to Tic32. Proc. Natl. Acad. Sci. USA 103: 16051–16056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ B., Hörtensteiner S. (2014). Mechanism and significance of chlorophyll breakdown. J. Plant Growth Regul. 33: 4–20. [Google Scholar]
- Christ B., Hauenstein M., Hörtensteiner S. (2016). A liquid chromatography-mass spectrometry platform for the analysis of phyllobilins, the major degradation products of chlorophyll in Arabidopsis thaliana. Plant J., http://dx.doi.org/10.1111/tpj.13253. [DOI] [PubMed] [Google Scholar]
- Christ B., Schelbert S., Aubry S., Süssenbacher I., Müller T., Kräutler B., Hörtensteiner S. (2012). MES16, a member of the methylesterase protein family, specifically demethylates fluorescent chlorophyll catabolites during chlorophyll breakdown in Arabidopsis. Plant Physiol. 158: 628–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ B., Süssenbacher I., Moser S., Bichsel N., Egert A., Müller T., Kräutler B., Hörtensteiner S. (2013). Cytochrome P450 CYP89A9 is involved in the formation of major chlorophyll catabolites during leaf senescence in Arabidopsis. Plant Cell 25: 1868–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro D.J., Gakhar L., Ramaswamy S. (2005). Rieske business: structure-function of Rieske non-heme oxygenases. Biochem. Biophys. Res. Commun. 338: 175–190. [DOI] [PubMed] [Google Scholar]
- Ginsburg S., Schellenberg M., Matile P. (1994). Cleavage of chlorophyll-porphyrin. Requirement for reduced ferredoxin and oxygen. Plant Physiol. 105: 545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J., Janick-Buckner D., Buckner B., Close P.S., Johal G.S. (2002). Light-dependent death of maize lls1 cells is mediated by mature chloroplasts. Plant Physiol. 130: 1894–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J., Wardzala E., Yang M., Reinbothe S., Haller S., Pauli F. (2004). A small family of LLS1-related non-heme oxygenases in plants with an origin amongst oxygenic photosynthesizers. Plant Mol. Biol. 54: 39–54. [DOI] [PubMed] [Google Scholar]
- Hibino T., Waditee R., Araki E., Ishikawa H., Aoki K., Tanaka Y., Takabe T. (2002). Functional characterization of choline monooxygenase, an enzyme for betaine synthesis in plants. J. Biol. Chem. 277: 41352–41360. [DOI] [PubMed] [Google Scholar]
- Hori K., et al. (2014). Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 5: 3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörmann F., Küchler M., Sveshnikov D., Oppermann U., Li Y., Soll J. (2004). Tic32, an essential component in chloroplast biogenesis. J. Biol. Chem. 279: 34756–34762. [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S. (2013a). The pathway of chlorophyll degradation: catabolites, enzymes and pathway regulation. In Plastid Development in Leaves during Growth and Senescence, Biswal B., Krupinska K., eds (Dordrecht, The Netherlands: Springer; ), pp. 363–392. [Google Scholar]
- Hörtensteiner S. (2013b). Update on the biochemistry of chlorophyll breakdown. Plant Mol. Biol. 82: 505–517. [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S., Feller U. (2002). Nitrogen metabolism and remobilization during senescence. J. Exp. Bot. 53: 927–937. [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S., Kräutler B. (2011). Chlorophyll breakdown in higher plants. Biochim. Biophys. Acta 1807: 977–988. [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S., Vicentini F., Matile P. (1995). Chlorophyll breakdown in senescent cotyledons of rape, Brassica napus L.: enzymatic cleavage of phaeophorbide a in vitro. New Phytol. 129: 237–246. [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S., Wüthrich K.L., Matile P., Ongania K.-H., Kräutler B. (1998). The key step in chlorophyll breakdown in higher plants. Cleavage of pheophorbide a macrocycle by a monooxygenase. J. Biol. Chem. 273: 15335–15339. [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S., Rodoni S., Schellenberg M., Vicentini F., Nandi O.I., Qiu Y.-L., Matile P. (2000). Evolution of chlorophyll degradation: the significance of RCC reductase. Plant Biol. 2: 63–67. [Google Scholar]
- Jockusch S., Turro N.J., Banala S., Kräutler B. (2014). Photochemical studies of a fluorescent chlorophyll catabolite--source of bright blue fluorescence in plant tissue and efficient sensitizer of singlet oxygen. Photochem. Photobiol. Sci. 13: 407–411. [DOI] [PubMed] [Google Scholar]
- Kim C., Apel K. (2004). Substrate-dependent and organ-specific chloroplast protein import in planta. Plant Cell 16: 88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kräutler B. (2014). Phyllobilins--the abundant bilin-type tetrapyrrolic catabolites of the green plant pigment chlorophyll. Chem. Soc. Rev. 43: 6227–6238. [DOI] [PubMed] [Google Scholar]
- Kräutler B. (2016). Breakdown of chlorophyll in higher plants - Phyllobilins as abundant, yet hardly visible signs of ripening, senescence, and cell death. Angew. Chem. Int. Ed. Engl. 55: 4882–4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kräutler B., Hörtensteiner S. (2014). Chlorophyll breakdown: chemistry, biochemistry and biology. In Handbook of Porphyrin Science: Chlorophyll, Photosynthesis and Bio-inspired Energy, Ferreira G.C., Kadish K.M., Smith K.M., Guilard R., eds (Singapore: World Scientific Publishing; ), pp. 117–185. [Google Scholar]
- Kräutler B., Jaun B., Bortlik K.-H., Schellenberg M., Matile P. (1991). On the enigma of chlorophyll degradation: the constitution of a secoporphinoid catabolite. Angew. Chem. Int. Ed. Engl. 30: 1315–1318. [Google Scholar]
- Küchler M., Decker S., Hörmann F., Soll J., Heins L. (2002). Protein import into chloroplasts involves redox-regulated proteins. EMBO J. 21: 6136–6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matile P., Schellenberg M., Peisker C. (1992). Production and release of a chlorophyll catabolite in isolated senescent chloroplasts. Planta 187: 230–235. [DOI] [PubMed] [Google Scholar]
- Matile P., Ginsburg S., Schellenberg M., Thomas H. (1988). Catabolites of chlorophyll in senescing barley leaves are localized in the vacuoles of mesophyll cells. Proc. Natl. Acad. Sci. USA 85: 9529–9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A., Eskandari S., Grallath S., Rentsch D. (2006). AtGAT1, a high affinity transporter for γ-aminobutyric acid in Arabidopsis thaliana. J. Biol. Chem. 281: 7197–7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser D., Matile P. (1997). Chlorophyll breakdown in ripening fruits of Capsicum annuum. J. Plant Physiol. 150: 759–761. [Google Scholar]
- Moser S., Müller T., Holzinger A., Lütz C., Jockusch S., Turro N.J., Kräutler B. (2009). Fluorescent chlorophyll catabolites in bananas light up blue halos of cell death. Proc. Natl. Acad. Sci. USA 106: 15538–15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlecker W., Ongania K.-H., Kräutler B., Matile P., Hörtensteiner S. (1997). Tracking down chlorophyll breakdown in plants: elucidation of the constitution of a ‘fluorescent’ chlorophyll catabolite. Angew. Chem. Int. Ed. Engl. 36: 401–404. [Google Scholar]
- Mühlecker W., Kräutler B., Moser D., Matile P., Hörtensteiner S. (2000). Breakdown of chlorophyll: a fluorescent chlorophyll catabolite from sweet pepper (Capsicum annuum). Helv. Chim. Acta 83: 278–286. [Google Scholar]
- Müller T., Ulrich M., Ongania K.H., Kräutler B. (2007). Colorless tetrapyrrolic chlorophyll catabolites found in ripening fruit are effective antioxidants. Angew. Chem. Int. Ed. Engl. 46: 8699–8702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhuber M., Berghold J., Breuker K., Hörtensteiner S., Kräutler B. (2003). Breakdown of chlorophyll: a nonenzymatic reaction accounts for the formation of the colorless “nonfluorescent” chlorophyll catabolites. Proc. Natl. Acad. Sci. USA 100: 6910–6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinská A., Tanner G., Anders I., Roca M., Hörtensteiner S. (2003). Chlorophyll breakdown: pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc. Natl. Acad. Sci. USA 100: 15259–15264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinská A., Anders I., Aubry S., Schenk N., Tapernoux-Lüthi E., Müller T., Kräutler B., Hörtensteiner S. (2007). In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 19: 369–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinská A., Tanner G., Aubry S., Anders I., Moser S., Müller T., Ongania K.-H., Kräutler B., Youn J.-Y., Liljegren S.J., Hörtensteiner S. (2005). Chlorophyll breakdown in senescent Arabidopsis leaves. Characterization of chlorophyll catabolites and of chlorophyll catabolic enzymes involved in the degreening reaction. Plant Physiol. 139: 52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinasabapathi B., Burnet M., Russell B.L., Gage D.A., Liao P.C., Nye G.J., Scott P., Golbeck J.H., Hanson A.D. (1997). Choline monooxygenase, an unusual iron-sulfur enzyme catalyzing the first step of glycine betaine synthesis in plants: prosthetic group characterization and cDNA cloning. Proc. Natl. Acad. Sci. USA 94: 3454–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodoni S., Mühlecker W., Anderl M., Kräutler B., Moser D., Thomas H., Matile P., Hörtensteiner S. (1997). Chlorophyll breakdown in senescent chloroplasts. Cleavage of pheophorbide a in two enzymic steps. Plant Physiol. 115: 669–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y., Schelbert S., Park S.-Y., Han S.-H., Lee B.-D., Andrès C.B., Kessler F., Hörtensteiner S., Paek N.-C. (2012). STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis. Plant Cell 24: 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelbert S., Aubry S., Burla B., Agne B., Kessler F., Krupinska K., Hörtensteiner S. (2009). Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 21: 767–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg M., Matile P., Thomas H. (1993). Production of a presumptive chlorophyll catabolite in vitro: requirement for reduced ferredoxin. Planta 191: 417–420. [Google Scholar]
- Scherl M., Müller T., Kreutz C.R., Huber R.G., Zass E., Liedl K.R., Kräutler B. (2016). Chlorophyll catabolites in fall leaves of the wych elm tree present a novel glyosylation motif. Chemistry 22: 9498–9503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheumann V., Klement H., Helfrich M., Öster U., Schoch S., Rüdiger W. (1999). Protochlorophyllide b does not occur in barley etioplasts. FEBS Lett. 445: 445–448. [DOI] [PubMed] [Google Scholar]
- Schuler M.A. (1996). Plant cytochrome P450 monooxygenases. Crit. Rev. Plant Sci. 15: 235–284. [Google Scholar]
- Schuler M.A., Duan H., Bilgin M., Ali S. (2006). Arabidopsis cytochrome P450s through the looking glass: a window on plant biochemistry. Phytochem. Rev. 5: 205–237. [Google Scholar]
- Sessions A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengel A., Soll J., Bölter B. (2007). Protein import into chloroplasts: new aspects of a well-known topic. Biol. Chem. 388: 765–772. [DOI] [PubMed] [Google Scholar]
- Stengel A., Benz P., Balsera M., Soll J., Bölter B. (2008). TIC62 redox-regulated translocon composition and dynamics. J. Biol. Chem. 283: 6656–6667. [DOI] [PubMed] [Google Scholar]
- Stengel A., Benz J.P., Buchanan B.B., Soll J., Bölter B. (2009). Preprotein import into chloroplasts via the Toc and Tic complexes is regulated by redox signals in Pisum sativum. Mol. Plant 2: 1181–1197. [DOI] [PubMed] [Google Scholar]
- Süssenbacher I., Christ B., Hörtensteiner S., Kräutler B. (2015). Hydroxymethylated phyllobilins in senescent Arabidopsis thaliana leaves - sign of a puzzling intermezzo of chlorophyll breakdown. Chemistry 21: 11664–11670. [DOI] [PubMed] [Google Scholar]
- Tanaka A., Ito H., Tanaka R., Tanaka N.K., Yoshida K., Okada K. (1998). Chlorophyll a oxygenase (CAO) is involved in chlorophyll b formation from chlorophyll a. Proc. Natl. Acad. Sci. USA 95: 12719–12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnády G.E., Simon I. (2001). The HMMTOP transmembrane topology prediction server. Bioinformatics 17: 849–850. [DOI] [PubMed] [Google Scholar]
- Wüthrich K.L., Bovet L., Hunziker P.E., Donnison I.S., Hörtensteiner S. (2000). Molecular cloning, functional expression and characterisation of RCC reductase involved in chlorophyll catabolism. Plant J. 21: 189–198. [DOI] [PubMed] [Google Scholar]
- Xodo L.E., Rapozzi V., Zacchigna M., Drioli S., Zorzet S. (2012). The chlorophyll catabolite pheophorbide a as a photosensitizer for the photodynamic therapy. Curr. Med. Chem. 19: 799–807. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.