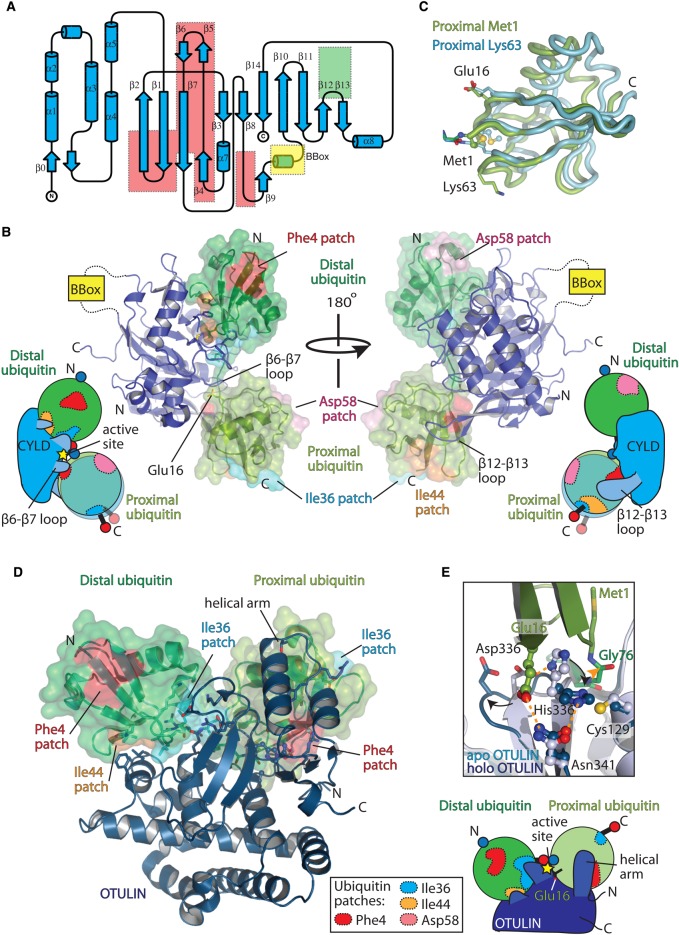
Figure 6. Molecular basis of Met1 disassembly (‘erasers’).
(A) Toplogy representation (generated by TopDraw [148]) of CYLD USP domain (PDB ID: 2VHF). Regions of the USP domain that are absent from CYLD compared with other USP domains are enclosed in a red box, whereas, the β12–β13 region that contains an insertion unique to CYLD is enclosed in a green box. The region that is replaced by an inserted B-box domain is shown in yellow. (B) Structure of CYLD ΔB-box (blue) bound to Met1-linked diubiquitin (green surface) (PDB ID: 3WXE) is shown in two different orientations. Cartoon representation showing the binding of Lys63 and Met1 diubiquitin is shown for each orientation. The different relative positions of the proximal ubiquitin moieties for Met1- and Lys63-linked diubiquitin is shown, as are the β6–β7 and β12–β13 loops. (C) Superimposition of the Met1 (green) and Lys63 (blue) proximal ubiquitin moieties from the CYLD diubiquitin structures (PDB ID: 3WXE and 3WXG, respectively), highlighting the positioning of the Met1 amino-terminus and Lys63 ε-NH2 side chain that forms the peptide/isopeptide bond, respectively. (D) Structure of OTULIN (blue) bound to Met1-linked diubiquitin (green) (PDB ID: 3ZNZ). Bottom right, schematic of the OTULIN interaction with Met1-linked diubiquitin. (E) Zoom-in of the catalytic site of apo OTULIN (light blue, PDB ID: 3ZNV) and holo OTULIN bound to Met1-linked diubiquitin (blue, PDB ID: 3ZNZ), showing the changes that occur within the active site upon Met1-linked diubiquitin binding. Hydrogen bonds between the catalytic triad residues (His336 and Asn341) and Glu16 are shown as orange dashes. The carbonyl of Met1 diubiquitin that is attacked by the nucleophilic catalytic cysteine (Cys129) is shown by an orange triangle.