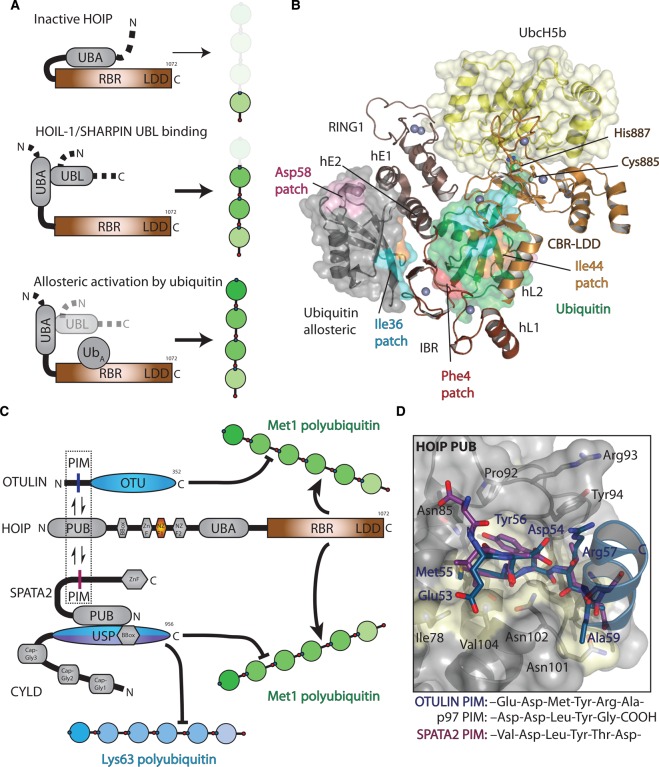
Figure 7. Regulation of the Met1 signal.
(A) Top, HOIP is autoinhibited through suspected binding of its UBA domain to the RBR domain, preventing Met1-linked polyubiquitin synthesis. Middle, binding of either HOIL-1 or SHARPIN UBL domains to the HOIP UBA domain releases autoinhibition and activates HOIP, allowing generation of Met1-linked polyubiquitin chains. Bottom, in addition to HOIL-1 or SHARPIN binding, the HOIP RBR domain contains an allosteric ubiquitin-binding site (UbA) that can also activate Met1 polyubiquitin formation. (B) Structure of the HOIP RBR bound to E2 (UbcH5b)∼ubiquitin (yellow and green surface) and non-covalent ubiquitin (ubiquitin allosteric, grey surface; PDB ID: 5DEV). Different hydrophobic patches are shown on both ubiquitin surfaces, showing the extensive interactions between the RBR and bound ubiquitin. (C) The activity of LUBAC is further regulated through DUB (OTULIN and CYLD) binding to the HOIP PUB domain. OTULIN contains an internal PIM that allows it to bind to the HOIP PUB domain. However, CYLD does not contain a PIM but is bound to SPATA2, which contains a PUB domain that specifically recognises the CYLD USP domain. SPATA2 also contains an internal PIM that allows it to bind to HOIP in an identical way as OTULIN. (D) Structure of the OTULIN PIM (PDB ID: 4OYK, blue) bound to the HOIP PUB domain (grey surface with interacting residues coloured yellow) and the SPATA2 PIM (PDB ID: 5LJN, purple). For clarity, only the OTULIN residues are annotated in blue. The PIM sequences that are able to bind the HOIP PUB domain are shown below for OTULIN, p97, and SPATA2.