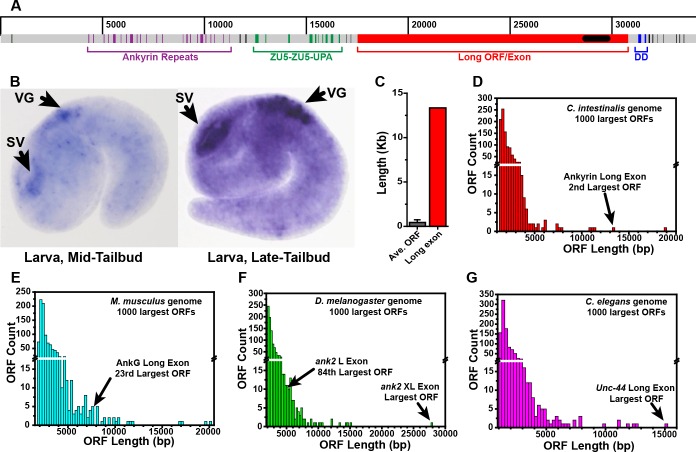
Fig 2. Ciona intestinalis ankyrin has a long exon comparable to mouse, fly and worm that can encode giant isoform(s).
(A) A map of the Ciona intestinalis ankyrin locus showing the position of exons (color coded by domain) comprising an ankyrin ortholog gene prediction relative to the position of a giant ORF (red) that could represent a long exon. (B) In situ hybridization of mid (left) and late (right) tail bud stage larvae with a probe corresponding to an EST cluster from within the potential long exon (black box in A) shows expression in the sensory vesicle (SV) and the visceral ganglion (VG), which together comprise the bulk of the larval nervous system. Pictures are publically available from the Aniseed database (Ascidian Network for In Situ Expression and Embryological Data; http://www.aniseed.cnrs.fr/) where additional pictures and methods can be found (gene prediction KH.C1.943). (C) Comparison of the Ciona ankyrin long exon size with that of the average size of all ORFs (defined as > 300 nucleotides without a stop codon) in the Ciona genome draft. (D) Length distribution for the largest 1,000 ORFs in the Ciona genome in bins of 250 bp, reveals that the Ciona ankyrin long exons sits in the 2nd largest ORF in the genome. (E-G) Similar length distribution graphs for the largest 1,000 ORFs in mouse, Drosophila melanogaster and C. elegans with the rank of long ankyring exons indicated.