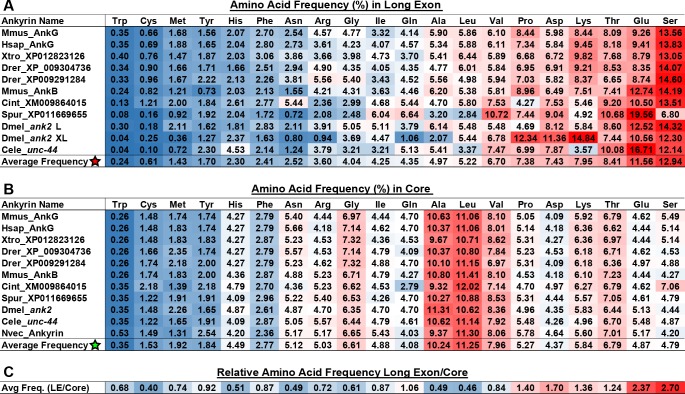
Fig 5. Amino acid composition of ankyrin long exon polypeptides.
(A) Amino acid compositions for 11 long exon-encoded polypeptides from 10 bilaterian ankyrin genes. Gene names are given at the left margin, and frequencies are given as percent composition for each amino acid. Frequencies are color coded with a blue (low) to red (high) and amino acids are arranged left to right in ascending order of average frequency. (B) An identical amino acid composition analysis as shown in (A) is given for the core ankyrin motifs (24 ankyrin repeats + the ZU5-ZU5-UPA cassette). Note the very different amino acid enrichment profiles (red highlights). For instance, Ala and Leu replace Glu and Ser as the top two most frequent residues. Nematostella ankyrin was included in the core analysis to demonstrate that the composition bias of the core has not changed since the divergence of cnidarians and bilaterians, likely soon after emergence of the first ankyrins. (C) Fold enrichment for each amino acid in long exon vs. core polypeptides, color coded blue (depleted) to red (enriched). The top 6 long exon amino acids are all enriched relative to the core with a > 2-fold enrichment for Glu and Ser. Fold enrichment was calculated for sequence sets shown in A and B by dividing the average amino acid frequency encoded in long exons (A, asterisk) by the average amino acid frequency found in the core (B, asterisk).