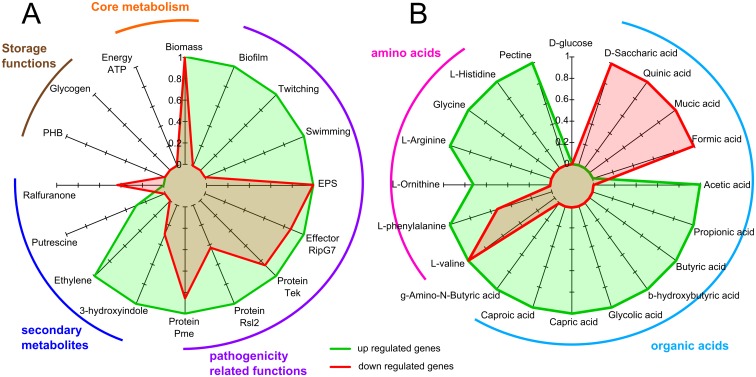
Fig 2. Predictions of phenotypes and catabolic pathways affected by EfpR-dependent DEGs (EfpR-DEGs) using flux balance analysis.
Simulations were performed using the genome-scale metabolic network of R. solanacearum. (A) Probability of the phenotypes to be affected by EfpR-DEGs ranking from 0 (not affected in any environments tested) to 1 (affected in all environments tested). (B) Probability of the catabolic capacities to be affected by EfpR-DEGs. Green, up-regulated genes; red, down regulated genes. Pme: pectin methyl esterase secretion; Rsl2: mannose-binding lectin, cell attachment; EPS: exopolysaccharide production; PHB: polyhydroxybutyrate.