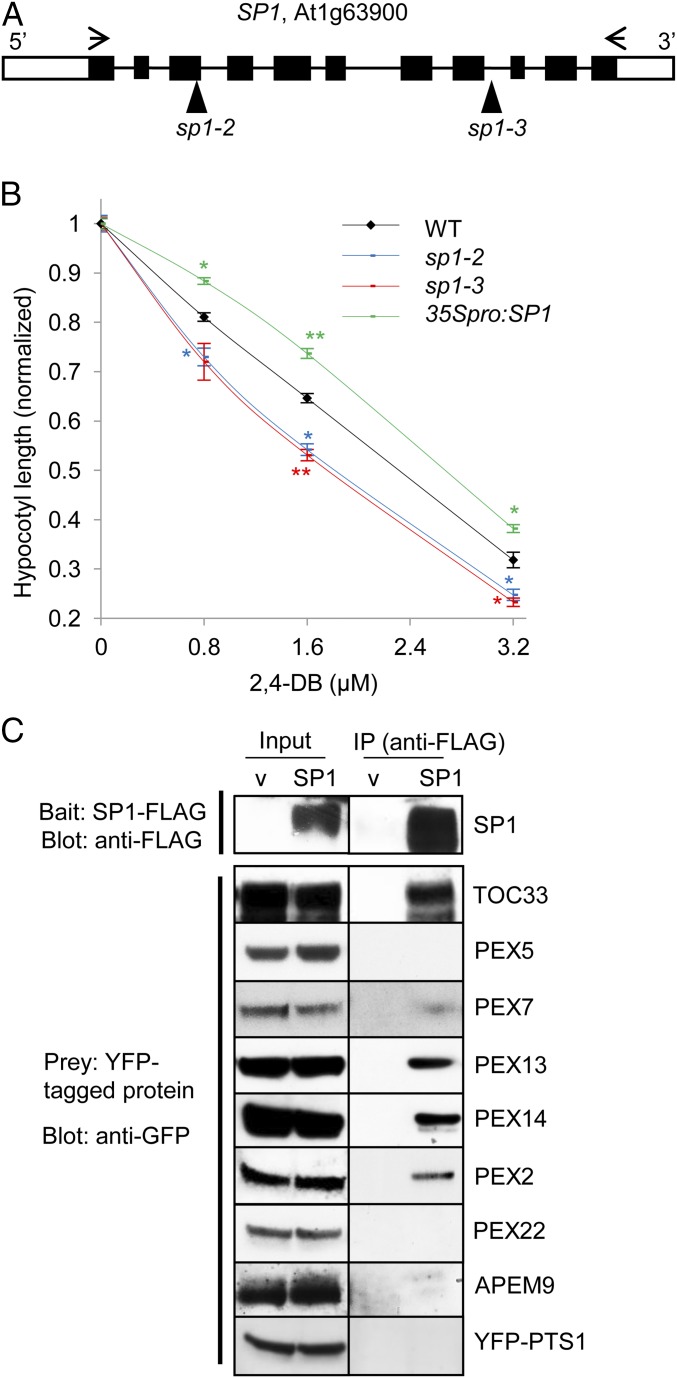
Fig. 2.
SP1 affects peroxisome function and interacts with several peroxins. (A) SP1 gene structure. The open box represents the UTR; the black boxes represent exons; the solid line represents the intron. T-DNA insertion sites in the two sp1 mutants are indicated. Arrows indicate the primers used to amplify full-length cDNA. (B) 2,4-DB response assays. Seedlings were grown on Murashige and Skoog (MS) medium supplemented with 0.5% sucrose and various concentrations of 2,4-DB. The average hypocotyl length of 7-d-old dark-grown seedlings is shown. All data on 2,4-DB are normalized to the data obtained from plants grown on 0.5% sucrose medium with no 2,4-DB. Data are shown as mean ± SEM; n = 3 (>30 seedlings were used for each biological replicate). **P < 0.01; *P < 0.05 from wild type. (C) Co-IP of PEX proteins with SP1. YFP-tagged candidate substrate proteins were coexpressed in tobacco leaves with either SP1-FLAG or empty vector (v). The top panel is a representative blot showing the level of SP1-FLAG in co-IP experiments performed for different YFP-tagged proteins.