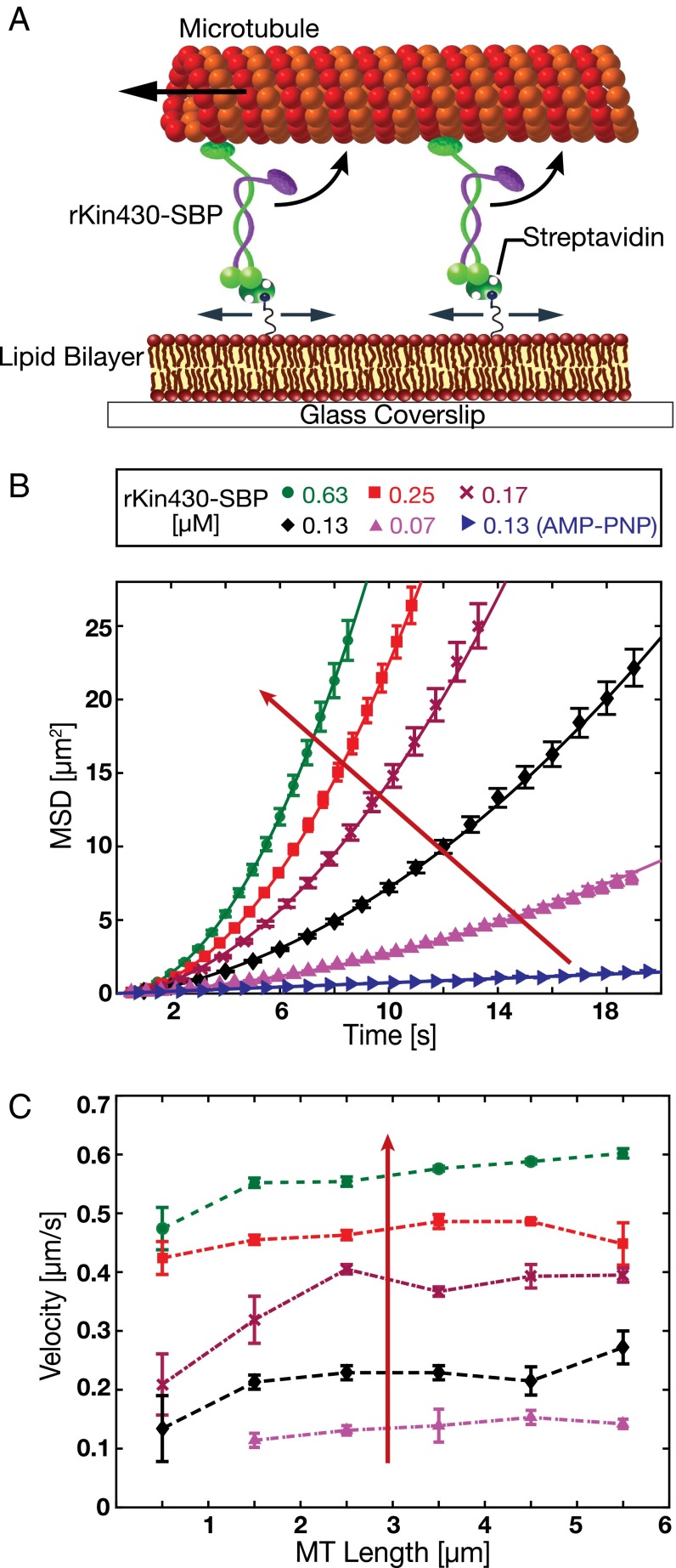
Fig. 2.
In vitro reconstitution of a membrane-anchored gliding motility assay. (A) Schematic drawing (not drawn to scale) of the experimental setup: truncated rat kinesin-1 with streptavidin-binding peptide tag (rKin430-SBP) is attached, via streptavidin, to a biotinylated SLB. The motors diffusively anchored on the SLB propel the microtubules. (B) Representative ensemble MSD data for the center positions of the microtubules (mean ± SEM; n ≥ 40 microtubules) at different motor concentrations in 1 mM ATP and 1 mM AMP-PNP (only for 0.13 µM rKin430-SBP). The red arrow indicates increasing motor concentration. To calculate the linear translocation components (i.e., the microtubule velocities νMT) the data were fit by Eq. 1. (C) Ensemble-averaged microtubule gliding velocities for different microtubule lengths, binned into 1-µm intervals, at different motor concentrations (mean ± 95% confidence interval; n ≥ 15 microtubules for each data point).