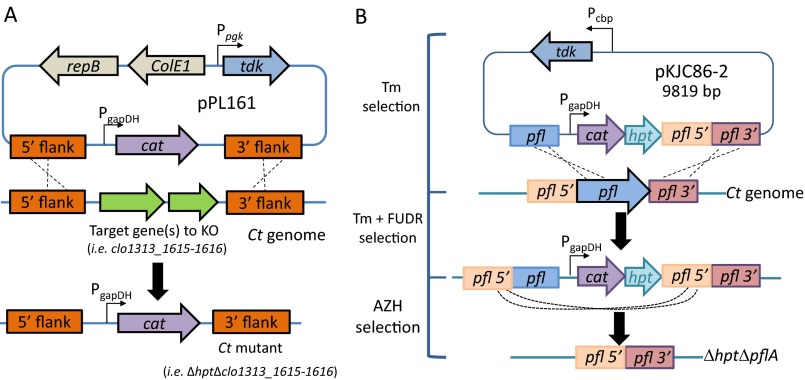
Fig. S2.
Schematics of constructs to knock out the gene(s) of interest. (A) Plasmid design to knock out putative PFOR individually in the C. thermocellum (Ct) genome via homologous recombination. The design replaced the target gene(s) with the expression of the thiamphenicol resistance gene (cat) on the genome for selection. Individual putative PFOR deletion mutants created are ∆hpt∆clo1313_1615-1616, ∆hpt∆clo1313_0673, ∆hpt∆clo1313_0382-0385, ∆hpt∆clo1313_0020-0023, and ∆hpt∆clo1313_1353-1356. (B) Schematics of markerless deletion of pflA gene from the C. thermocellum (Tm) genome. All mutants were generated in the ∆hpt background. All PCR products were validated by commercial Sanger sequencing. AZH, azahypoxanthin; ColE1, Escherichia coli replication origin; FUDR, 5-fluoro-2′deoxyuridine; Pcbp, cellobiose phosphorylase promoter; Ppgk, phosphoglycerate kinase promoter; repB, gram-positive replication origin; tdk, thymidine kinase.