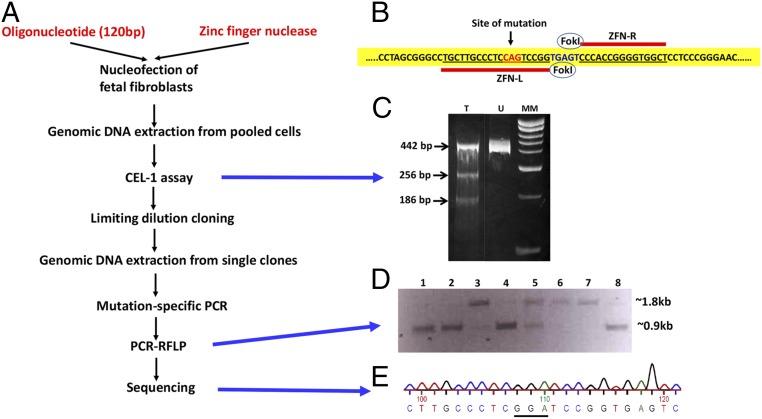
Fig. 1.
Development of bovine fetal fibroblasts containing the Q(‒5)G substitution in both alleles of CD18. (A) Schematic representation of ZFN-mediated gene editing of bovine ITGB2. (B) ZFN-mediated induction of the Q(‒5)G substitution. The pair of ZFNs were designed to cleave bovine ITGB2 in the vicinity of the codon targeted for editing (red fonts). Sequences bound by each ZFN are underlined in red. ITGB2 domain targeted for FokI-cleavage is in blue fonts. (C) Cel-1 assay. Lanes T and U show the digestion products from the ZFN-transfected and ZFN-untransfected cells, respectively. Lane MM represents molecular weight markers. (D) PCR-RFLP assay. The ∼1.8-kb and ∼0.9-kb bands represent the undigested and digested products, respectively. In this figure, lanes 1, 2, 4, and 8 show clones that are homozygous, whereas lane 5 shows a clone that is heterozygous for the mutation. Lanes 3, 6, and 7 show clones without any modification. (E) Sequencing analysis. Sequencing data shows the presence of glycine-encoding GGA codon instead of the glutamine-encoding CAG codon.