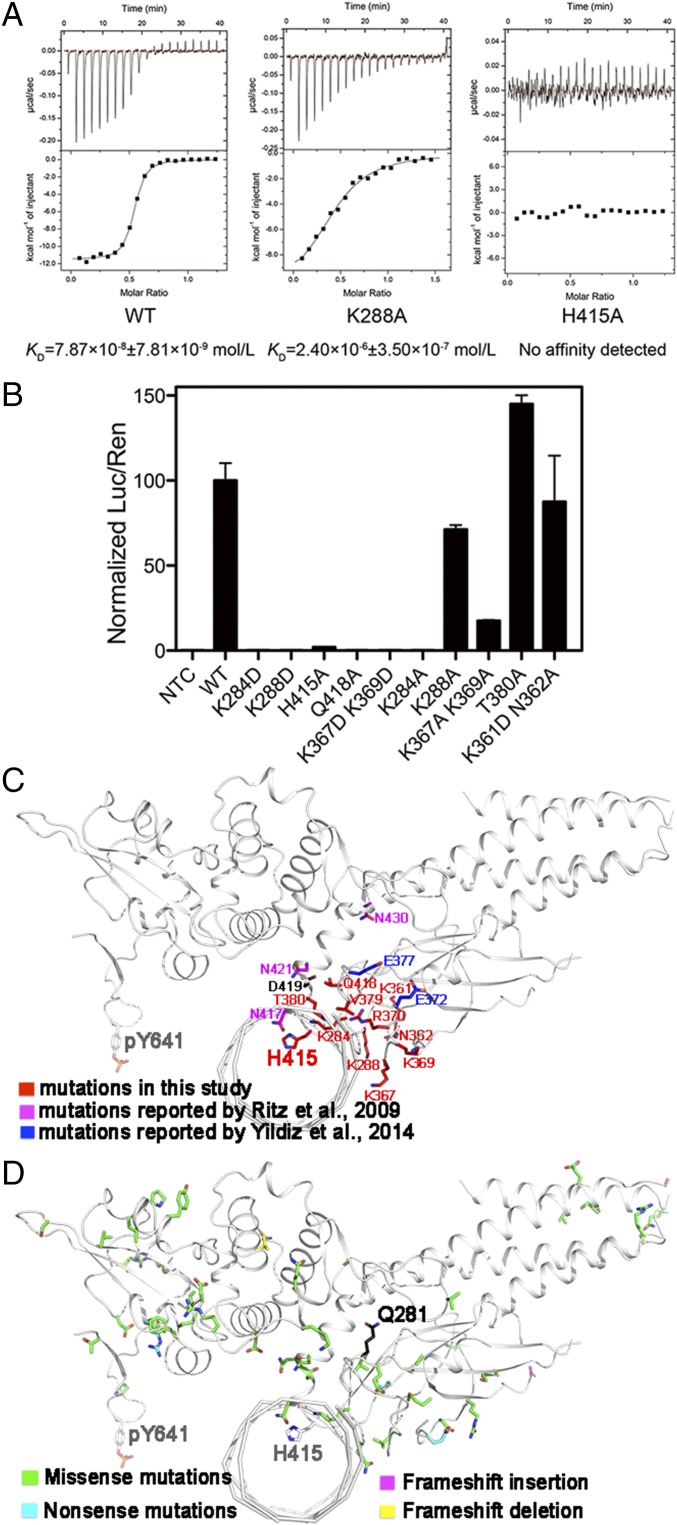
Fig. 4.
Identification of key residues for DNA binding by mutagenesis. (A) ITC measurements of affinities of STAT6CF-WT (Left), STAT6CF-K288A (Middle), and STAT6CF-H415A (Right) for N4 site DNA (CS4) are shown. (B) The ability of several mutants of STAT6FL to activate gene transcription was assessed. Mean ratio luciferase/renilla light units activities are shown for triplicate samples. Normalized results are presented as percent activity relative to the activity in cells transfected with STAT6FL-WT. (C) The residues (in red) mentioned above for mutagenesis, mutations reported by Ritz et al. (in magenta) (26) and Yildiz et al. (in blue) (22) were mapped on a protomer of STAT6CF-N4 structure (in front view). The key residue Y641 is also shown. (D) Unique mutations from a large-scale cancer genomics dataset analysis in cBioPortal (www.cbioportal.org) web tool are mapped on a protomer of STAT6CF-N4 structure.