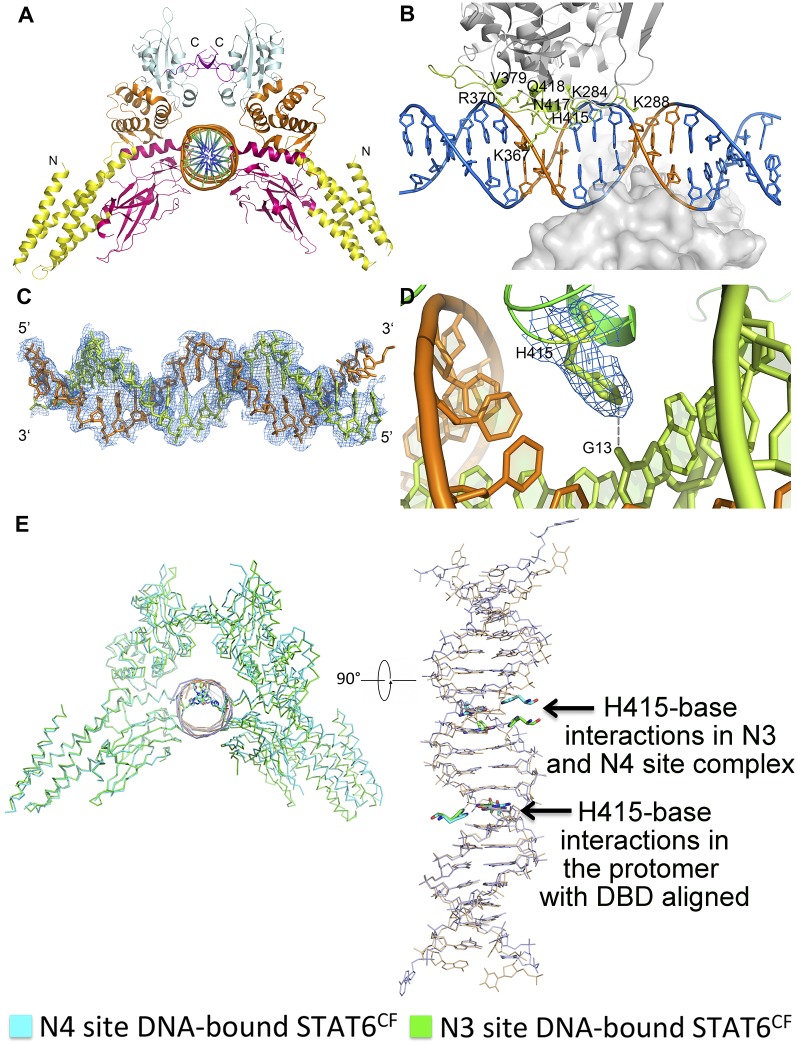
Fig. S5.
Crystal structure of STAT6CF-N3 complex and its comparison with STAT6CF-N4 complex structure. (A) Cartoon diagram of the STAT6CF-N3 complex. Colors of each domain are the same as in Fig. 1B. (B) Drawing depicting details of the STAT6CF-N3 interface. The side chains of residues (A chain) donating hydrogen bonds are shown by sticks in lemon and hydrogen bonds are shown in gray dash. The conserved palindromic bases (TTC/GAA) are shown in orange. (C) A simulated annealing (SA) omit electron density map (2Fo-Fc), contoured at 1.0σ, of N3 site DNA in STAT6CF-N3 complex. (D) Electron density map (2Fo-Fc), contoured at 1.0σ, of H415 at DNA binding domain of STAT6. Hydrogen bond formed by H415 and G13 of DNA chain is shown in gray dash. (E) Conformational changes between STAT6CF-N3 and N4 site DNA complexes. The DNA binding domains are aligned together in one protomer of the STAT6CF dimer and the difference in the movement of H415-base interactions in the DNA binding domains in another protomer of the STAT6CF dimer is shown (in bottom view).