Single-molecule tracking and spatial stochastic modeling reveal receptor dimer stabilization by nanoscale confinement zones.
Keywords: Cytokine receptor signaling, type I interferon receptor, plasma membrane organization, hop diffusion, protein-protein interaction, single molecule tracking, spatial stochastic modeling
Abstract
The interaction dynamics of signaling complexes is emerging as a key determinant that regulates the specificity of cellular responses. We present a combined experimental and computational study that quantifies the consequences of plasma membrane microcompartmentalization for the dynamics of type I interferon receptor complexes. By using long-term dual-color quantum dot (QD) tracking, we found that the lifetime of individual ligand-induced receptor heterodimers depends on the integrity of the membrane skeleton (MSK), which also proved important for efficient downstream signaling. By pair correlation tracking and localization microscopy as well as by fast QD tracking, we identified a secondary confinement within ~300-nm-sized zones. A quantitative spatial stochastic diffusion-reaction model, entirely parameterized on the basis of experimental data, predicts that transient receptor confinement by the MSK meshwork allows for rapid reassociation of dissociated receptor dimers. Moreover, the experimentally observed apparent stabilization of receptor dimers in the plasma membrane was reproduced by simulations of a refined, hierarchical compartment model. Our simulations further revealed that the two-dimensional association rate constant is a key parameter for controlling the extent of MSK-mediated stabilization of protein complexes, thus ensuring the specificity of this effect. Together, experimental evidence and simulations support the hypothesis that passive receptor confinement by MSK-based microcompartmentalization promotes maintenance of signaling complexes in the plasma membrane.
INTRODUCTION
Signal transduction across the plasma membrane (PM) frequently involves the association of transmembrane receptors to dimeric or oligomeric complexes. These complexes are formed by reversible, noncovalent interactions and thus continuously associate and dissociate. Nanoscale hierarchical organization of the PM caused by the cortical actin meshwork [membrane skeleton (MSK)], membrane microdomains (lipid rafts), and protein-protein interactions (1–8) has been speculated to play an important role in the assembly and dynamics of signaling complexes (9–14). In particular, dimerization of receptor tyrosine kinases and related receptors has been suspected of being highly regulated by PM compartmentalization (15–17). However, the function of transient confinement by nanoscale membrane domains in sustaining individual, functional signaling complexes has remained largely speculative (18).
Here, we have explored the assembly and dynamics of the type I interferon (IFN) receptor by single quantum dot (QD) tracking and localization microscopy (TALM). The IFN receptor comprises two subunits, IFNAR1 and IFNAR2, which are expressed at low cell surface densities (typically a few 100 copies per cell) (19). IFNAR1 and IFNAR2 are dimerized by simultaneous interaction with IFN via independent binding epitopes (19–21). IFNAR2 binds IFNα2 with an equilibrium dissociation constant of ~5 nM and a complex lifetime of ~100 s (22), whereas the affinity toward IFNAR1 is three orders of magnitude lower, and the complex dissociates about 100-fold faster (20, 23). Different members of the IFN family bind IFNAR1 with distinct binding affinities and kinetics, which has been correlated with their differential cellular activities (24, 25). By exploiting recent progress in nanoparticle monofunctionalization and receptor cell surface labeling (26), we unraveled here the diffusion and interaction dynamics of individual receptor dimers in the PM of living cells by single QD tracking (27–32), enabling long-term observation of individual IFNAR signaling complexes with very high spatial and temporal resolution (Fig. 1A). On the basis of spatiotemporal correlation analyses of single complex trajectories, we identified transient dissociation and reassociation of complexes within MSK nanostructures. Integrating our comprehensive experimental data with quantitative spatial stochastic models suggests that hierarchical, two-tiered PM compartmentalization governs efficient reassociation of receptor dimers, implicating a sensitive interplay of MSK-based diffusion barriers and receptor dimerization dynamics.
Fig. 1. Spatiotemporal dynamics of IFNAR assembly in the PM.
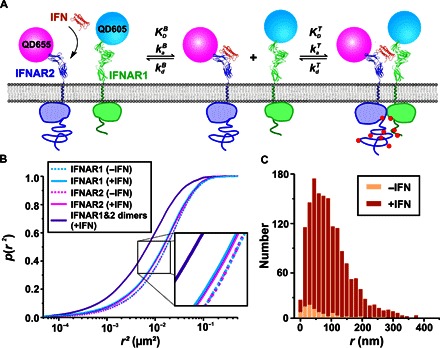
(A) IFNAR1 and IFNAR2 stably expressed in U5A cells at physiological levels were labeled orthogonally with monofunctional QD605 and QD655. Rate constants involved in a two-step receptor dimerization mechanism, which can be assumed owing to the highly asymmetric properties of IFN interaction with the two receptor subunits: After IFN capturing from the solution by the high-affinity subunit IFNAR2, IFNAR1 is recruited within the membrane to form the ternary signaling complex. The dynamic equilibrium between binary and ternary complexes in the PM is described by the 2D rate constants and as well as the 2D equilibrium dissociation constant . (B) Cumulative probability plot of QD-labeled IFNAR1 and IFNAR2 square displacements in the absence (dashed lines) and in the presence (solid lines) of 50 nM IFNα2 as obtained from >10,000 individual displacements. The square displacements were determined for a lag time of 160 ms (five frames). The IFNα2 concentration ensured saturation of all IFNAR2 at the cell surface. For comparison, the square displacements observed for IFNAR1/IFNAR2 dimers are included (violet line). Diffusion constants obtained by two-component mean square displacement (MSD) analyses are provided in table S1. (C) Histogram of the displacements of IFNAR1-IFNAR2 dimers identified by colocomotion analysis in the absence and in the presence of IFNα2 (N = 81 without IFNα2 versus N = 1492 with IFNα2) for a lag time of 160 ms.
RESULTS
Dimerization of IFNAR1 and IFNAR2 is induced by IFN
IFNAR dimerization and the lifetime of receptor dimers at the PM were explored by single-molecule localization microscopy using QD labeling, ensuring high photostability and long-term imaging with the highest spatial and temporal resolution (27). For site-specific, orthogonal labeling with QDs, IFNAR1 and IFNAR2 were fused to the HaloTag and the SNAP tag, respectively, and stably expressed in IFNAR2-deficient U5A cells at physiological levels (~5 to 10 copies/μm2). Full functional integrity of this complemented cell line with respect to IFN signaling was recently demonstrated (33), making it a viable model for our studies. On the basis of sequential, tag-specific biotinylation, IFNAR1 and IFNAR2 were labeled with monobiotinylated QDs via bivalent streptavidin (bSAV) (Fig. 1A) (26), thus excluding bias by multivalent interactions. A typical labeling degree of ~16% with high orthogonality and negligible background labeling was achieved (movies S1 and S2 and fig. S4). Statistical analysis of displacements (within five frames, that is, 160 ms), obtained from more than 10,000 individual displacements, revealed highly similar diffusion kinetics of IFNAR1 and IFNAR2 (Fig. 1B). Upon the addition of IFNα2, a small yet significant reduction in the average diffusion could be detected, suggesting ligand-induced receptor dimerization (Fig. 1B). Single-molecule colocomotion analysis based on tracking of colocalized molecules over multiple steps revealed that only after the addition of the ligand could a substantial amount of IFNAR1/IFNAR2 dimers be detected (Fig. 1C and movie S3). The mobility of these receptor dimers was substantially decreased, as compared to the unliganded receptor subunits (Fig. 1B and table S1). The association and dissociation of individual receptor dimers could be discerned in the presence of IFNα2 (movie S4 and Fig. 2, A and B), corroborating the model of reversible receptor dimerization, as depicted in Fig. 1A. From the relative number of colocomotion trajectories after IFNα2 stimulation (6.6%) in combination with the estimated QD labeling degree of ~16%, we infer that ~40% of receptors were recruited into ternary complexes. In comparison, ~2% receptor dimers were observed in the absence of ligand, which is characterized by very low mobility and can be ascribed to nonspecific background binding and/or aggregation of QD. These results highlight the efficient IFNα2-induced dimerization of IFNAR1 and IFNAR2 and the formation of a dynamic equilibrium between binary and ternary complexes in the PM.
Fig. 2. Dynamic equilibrium of receptor dimerization observed at the single-molecule level.
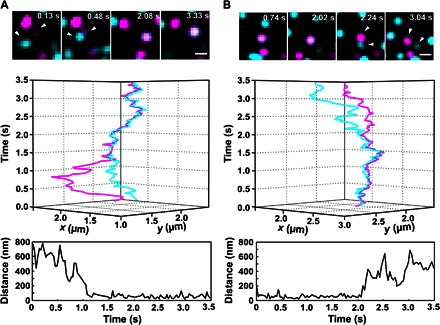
Assembly (A) and dissociation (B) of individual IFNAR1 (cyan) and IFNAR2 (magenta) heterodimers observed in the presence of IFNα2: time series of images (top), 3D trajectories (middle), and distances between receptors as a function of time (bottom). White arrowheads mark the dimerized receptors. Scale bars, 1 μm.
Lifetime and effector activation of receptor dimers depend on MSK integrity
Even though individual ternary complexes could be readily identified by colocomotion analysis, insufficient tracking fidelity due to QD blinking impeded the reliable quantification of lifetimes of individual complexes from the length of cotrajectories. We therefore devised a method to filter the data sets according to time-lapse particle correlation (fig. S5), yielding much longer and largely unbiased colocomotion trajectories. On the basis of this method, we analyzed the lifetime of several hundred individual receptor dimers, yielding an exponential distribution with an average complex lifetime τ = 15.2 ± 1.7 s (Fig. 3, A and B). In contrast, the lifetime of ternary complexes in artificial membranes in vitro was found to be ~3 s (34), that is, fivefold lower than that of ternary complexes in the PM.
Fig. 3. Lifetime of receptor dimers and IFN signaling activity depend on MSK integrity.
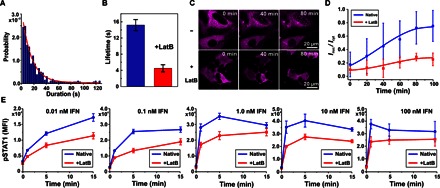
(A and B) Ternary complex lifetime in the native PM obtained from a colocomotion trajectory length histogram (390 individual tracked colocomotion events). An average lifetime of 15.2 ± 1.7 s (mean ± SD) was obtained by an exponential fit. (B) Complex lifetimes obtained from trajectory length histograms in the absence and in the presence of LatB. (C and D) IFNα2-stimulated translocation of STAT2 in the absence and in the presence of LatB. (C) Translocation of STAT2-TagRFP stably expressed in HeLa cells, visualized by confocal laser scanning microscopy. (D) Time-dependent STAT2 translocation monitored by the ratio of fluorescence intensity of STAT2-TagRFP in the nucleus to that in the cytoplasm. Data are presented as means ± SD from seven cells connected by lines to guide the eye. (E) Kinetics of STAT1 phosphorylation at different IFNα2 concentrations in the absence and in the presence of LatB. HeLa cells were stimulated with the indicated concentrations of IFNα2, and pSTAT1 was quantified at various time points by phosphoflow cytometry. Data are means ± SE from four independent replicates.
Potentially, transient confinement of receptor dimers in submicroscopic MSK corrals (<100 nm in diameter) could be responsible for enhancing the probability that dissociated receptor subunits rapidly reassociate. To test this hypothesis, we investigated the role of the MSK in confining IFNAR1 and IFNAR2 and in the apparent lifetime of individual ternary complexes. Upon treatment of cells with Latrunculin B (LatB) to inhibit actin polymerization, thus destabilizing the MSK (35), approximately twofold increased diffusion constants for both IFNAR1 and IFNAR2 were detected (fig. S6 and table S1). Strikingly, increased receptor mobility upon LatB treatment was accompanied by a decrease in the stability of receptor dimers (Fig. 3B). Under these conditions, a lifetime of 4.4 ± 0.8 s was obtained, corroborating the fact that the MSK contributes to stabilizing signaling complexes.
Because the lifetime of signaling complexes may be critical for downstream signaling, we examined the role of MSK integrity in the activation of signal transducer and activator of transcription 1 (STAT1) and STAT2, which are hallmark effector proteins of IFN signaling (36, 37). STAT1 and STAT2 are phosphorylated by the IFN signaling complex to form, together with interferon regulatory factor 9, the transcription factor ISGF3, a key regulator of IFN-stimulated gene expression (36, 38). ISGF3 is formed upon phosphorylation of STAT1 and STAT2 by the IFN signaling complex, resulting in the nuclear translocation of these proteins (39). Functional studies were performed in HeLa cells, in which IFN downstream signaling is more robust than in the complemented U5A cell line, whereas both cell lines display similar receptor density, diffusion, and signaling dynamics (33, 40). In a first step, we quantified IFN-stimulated nuclear translocation of STAT2 by fluorescence microscopy in living cells. In native HeLa cells stably expressing STAT2 fused to TagRFP (STAT2-TagRFP), time-dependent translocation of STAT2 into the nucleus was observed after treatment with IFNα2 (Fig. 3C), yielding characteristic subnuclear accumulation (41). In contrast, a much weaker nuclear translocation was observed for the same experiment carried out after LatB treatment (Fig. 3, C and D). STAT2 translocation was still detectable after LatB treatment, confirming that IFN signaling was still possible in these cells but with decreased efficacy. To pinpoint the cause of reduced ISGF3 translocation, we explored how LatB treatment affects STAT phosphorylation. Kinetic analysis of STAT1 phosphorylation by phosphoflow cytometry confirmed the functional relevance of actin cytoskeleton integrity for IFN signaling (Fig. 3E). The concentration dependence and time dependence of STAT1 phosphorylation suggest that LatB treatment affects the rate of STAT1 phosphorylation. Because phosphorylated STAT1 is required for ISGF3 formation and nuclear translocation, this observation can explain delayed STAT2 translocation upon LatB treatment. These results suggest that the actin cytoskeleton is involved in the activation of IFN effector proteins, possibly as a scaffold of PM compartmentalization, contributing to stabilizing IFNAR1/IFNAR2 dimers. However, other cellular functions of actin polymerization (for example, in receptor endocytosis and trafficking) could also be responsible for this effect.
A spatial stochastic MSK model reproduces IFNAR diffusion properties
We therefore further explored our hypothesis that confinement by the nanoscale MSK meshwork, as proposed by the “picket fence” model (42), may promote rapid reassociation of the same receptor subunits. Because a diameter of <100 nm for the MSK confinement zone has been reported, the temporal and spatial resolution applied in the cotracking experiments did not allow us to resolve these events (3). To better understand the effect of MSK compartments on reassociation and the apparent stability of ternary complexes, we set up a high-resolution spatial stochastic computer model of receptor diffusion in the PM. Our model was parameterized using experimental molecular kinetics and diffusion data obtained with IFNAR reconstituted using artificial membranes (23, 34, 43) (see the Supplementary Materials for a detailed model description). An MSK-like compartmentalization was introduced by means of a triangular mesh with an average length scale of 70 nm (Fig. 4A and fig. S7). This length scale, corresponding to the mean square root of the compartment area, had been previously reported for HeLa cells (3, 6), which yielded very similar diffusion properties of IFNAR1 and IFNAR2 compared to U5A cells (33, 40). Hop diffusion between MSK compartments was specified by the associated hop probability pMSK, whereas the diffusion constants inside the corral were set at values obtained for the transmembrane proteins and the ternary complex reconstituted into artificial membranes (compare Table 1) (43, 44). As it turned out, this model could robustly reproduce the experimentally observed step length histograms in the absence and in the presence of LatB (Fig. 4B and fig. S8).
Fig. 4. Modeling of diffusion and interaction in the context of a stochastic MSK meshwork.

(A) Model of the MSK mesh with an average 70-nm length scale. Inset is a 2× magnification of the region highlighted by the rectangle. (B) Displacement probability density of the diffusing receptors (IFNAR1, circle; IFNAR2, triangle) in the PM of native cells (blue) and LatB-treated cells (red). Fitting the experimental data sets (dotted lines) with the MSK hop diffusion model yielded experimental hopping probability values summarized in Table 1. (C) Representative spatial stochastic simulation of an individual complex within MSK microcompartments, revealing transient dissociation and reassociation of receptor dimers (time lapse, 0.032 s; IFNAR1, cyan; binary IFNAR2/IFNα2 complex, magenta; ternary complex, blue). (D) Experimental lifetimes of ternary complexes measured in solid supported lipid bilayers (LB) and in the PM, compared to simulated ternary complex lifetimes based on receptor confinement by the MSK in native cells (blue) and in LatB-treated cells (red).
Table 1. Key experimental parameters and the associated constants derived by simulations.
Molecular diffusion constants of receptors in an artificial lipid bilayer Dmol (mean value ± SD) (43); diffusion constants in the PM Dcell obtained from 160 ms squared displacement analysis (mean values ± SD) (table S1); simulated hop probabilities pMSK; simulated apparent lifetime of complexes within the MSK τMSK; hop probability across SC barriers pSC. NA, not applicable.
| Cell status | Species |
Dmol (μm2/s) |
τmol (s) |
Dcell (μm2/s) |
τcell (s) |
pMSK |
τMSK (s) |
pSC |
τMSK+SC (s) |
| Native | Receptor subunits | 1.1 ± 0.2 | NA | 0.043 ± 0.002 | NA | 0.00095 | NA | 0.0025 | NA |
| Ternary complex | 0.7 ± 0.2 | 3 | 0.022 ± 0.001 | 15.2 ± 1.7 | 0.0006 | 9.4 | 0.0018 | 12.1 | |
| +LatB | Receptor subunits | 1.1 ± 0.2 | NA | 0.068 ± 0.004 | NA | 0.0036 | NA | NA | NA |
| Ternary complex | 0.7 ± 0.2 | 3 | 0.053 ± 0.002 | 4.4 ± 0.8 | 0.002 | 4.1 | NA | NA |
MSK-dependent complex reassociation events are predicted by the model
On the basis of the pMSK values obtained by fitting the experimental data (Table 1), we simulated the interaction dynamics of individual complexes using the molecular association and dissociation rate constants and , respectively (compare Fig. 1A), previously determined for IFNAR1 and IFNAR2 reconstituted into artificial membranes (23, 34, 43). The experimental was converted into an on-rate probability pon, assuming an association radius ron = 5 nm estimated from the structure of the ternary complex (21). In comparison with experiments, a pon value of 0.0068 was obtained, highlighting the fact that the recruitment of IFNAR1 by the IFNα2/IFNAR2 complex is far from being diffusion-controlled in homogeneous membranes. The average lifetime of the complex τmol was set to 3 s in accordance with , the IFNAR1 dissociation rate constant from the IFNAR2/IFNα2 complex (34). To determine the apparent complex lifetime in the MSK meshwork (τMSK), we initiated simulations with a single receptor dimer that was randomly placed inside the compartmentalized membrane domain. During diffusion in the MSK meshwork, the dissociation of the complex was stochastically initiated according to the average molecular lifetime τmol, and reassociation was allowed on the basis of pon. Although these simulations were carried out with a 5-μs time resolution and a <5-nm spatial resolution, the positions of the complex and the two receptor subunits were recorded every 0.032 s to mimic the experimental acquisition rate. A complex was considered fully dissociated once IFNAR2 and IFNAR1 were localized in non-neighboring MSK compartments (compare the Supplementary Materials). These simulations revealed substantial reassociation within individual MSK compartments for hopping probabilities of an intact MSK (movie S5, Fig. 4C, and fig. S10). A histogram of these apparent complex lifetimes obtained from 100 individual complexes is shown in Fig. 4D, yielding an approximately threefold increase in the average complex lifetime (τ = 9.4 s), as compared to the τmol of noncompartmentalized, artificial membranes. In contrast, only minor apparent stabilization of the ternary complex was observed in simulations with hopping rates corresponding to the presence of LatB (τ = 4.1 s; Fig. 4D), matching the experimentally observed complex lifetimes very well. The robustness of these results was corroborated by extensive control simulations with altered local diffusion properties of the receptor (fig. S11). Likewise, variation in ron did not yield significant changes in the complex stability, given that pon was adjusted according to the in vitro association kinetics (fig. S12).
Receptor diffusion is confined by hierachical, MSK-dependent microdomains
These results fostered our hypothesis that reassociation due to passive confinement by the MSK can increase the apparent stability of ternary complexes in the PM. However, our compartmentalization model could not match the experimentally observed ternary complex lifetimes in wild-type cells. We therefore explored in more detail the submicroscopic spatiotemporal organization of IFNAR1 and IFNAR2 in the PM. For this purpose, we used TALM (45), a robust method that we recently developed for identifying and characterizing transient confinement zones (40). Superresolution TALM images, reconstructed by localizing individual receptor subunits over a period of 25 s, revealed substantial local confinement of the receptors (Fig. 5A). To selectively quantify the dimension of the spatiotemporal organization of ternary complexes, we applied pair correlation TALM (pcTALM), which clearly identified transient confinement zones with an average diameter of 250 ± 30 nm (Fig. 5, B and C). Notably, these secondary confinement zones could not be detected in the presence of LatB (fig. S13), suggesting that these larger-scale microcompartments also depend on the cortical actin skeleton.
Fig. 5. Secondary compartmentalization of IFNAR complexes.
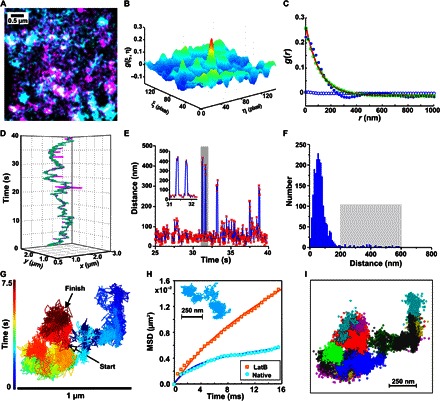
(A to C) Identification of secondary confinement zones by dual-color TALM and pcTALM. (A) TALM images of IFNAR1 (cyan) and IFNAR2 (magenta) in the presence of IFNα2. (B) Image cross-correlation and (C) pair correlation analyses. The pair correlation curve (blue dots) was deconvolved into the contributions of free (blue circles) and confined (green circles) diffusion components by fitting Eq. 3 (Materials and Methods). (D to F) Transient dissociation events observed within experimental colocomotion trajectories. (D) Localizations of IFNAR1 (cyan) and IFNAR2 (magenta) within an individual colocomotion trajectory. (E) IFNAR1-IFNAR2 distance map within an individual cotrajectory (red dots connected by a blue line) showing dissociation and reassociation events within secondary confinement dimensions. Two characteristic dissociation events highlighted by the gray bar are enlarged in the inset. (F) Histogram of IFNAR1-IFNAR2 distances yielding a significant fraction (2.3%) of distances of >200 nm, which correspond to transient dissociation events (highlighted by the gray rectangle). The total number of displacements N = 1560 was obtained from 24 trajectories yielding an average of 1.5 reassociation events per trajectory. (G and H) Detection of secondary confinement zones by fast QD tracking. (G) Trajectory of an individual particle covering 15,000 frames. Color coding according to the acquisition time indicated by the time bar. (H) MSD analysis of fast QD tracking in native cells and after the addition of LatB. A typical trajectory from native cells is shown in the inset. MSD data obtained from more than 1000 trajectories. (I) Identification of domains (depicted in different colors) by DBSCAN analysis.
Upon closer inspection of individual colocomotion trajectories, distances between IFNAR1 and IFNAR2 above the noise level were detected (Fig. 5, D to F, and movie S6), which is indicative of transient dissociation and reassociation events within these larger membrane compartments. Our analyses revealed an average of 1.7 reassociation events per trajectory with a significant intersubunit separation distance of >200 nm. We further characterized the properties of these secondary confinement zones by QD tracking with submillisecond time resolution (2000 frames per second) using a scientific complementary metal-oxide semiconductor (sCMOS) camera. A typical trajectory obtained at this 66-fold increased acquisition rate is shown in Fig. 5G, clearly exhibiting highly confined diffusion properties (Fig. 5H, fig. S14, and movie S7). Confinement zones were identified and quantified using a spatiotemporal cluster algorithm (Fig. 5I), yielding an average diameter of 270 nm and an average dwell time of 0.43 s (fig. S15). Characterization of the diffusion properties by MSD analysis confirmed nested confinement zones (Fig. 5H, fig. S14, and table S4). Upon the addition of LatB, secondary confinement was largely abrogated, which agrees with the pcTALM analysis very well.
Comprehensive spatial stochastic model reproduces experimental complex lifetimes
We therefore refined our membrane model to include additional microcompartmentalization as a secondary, passive transient confinement zone. To this end, we introduced hierarchical confinement with each secondary compartment (SC) set to be composed of a number of interconnected MSK compartments, as observed in the experiments (Fig. 6A and fig. S16). Inner (MSK-based) and outer (SC-based) barriers were assigned distinct hop probabilities (pMSK and pSC, respectively) according to the experimentally observed diffusion properties (fig. S17). On the basis of this refined spatial stochastic diffusion model, we simulated the dissociation of ternary complexes (compare the Supplementary Materials). By applying this two-tiered membrane compartmentalization model, a further increase of the apparent complex stability was obtained (τ = 12.1 s), more closely matching the experimentally observed complex stability (Fig. 6B). On the basis of this model, we reassessed our trajectory length analysis by terminating cotrajectories at an intersubunit distance threshold of 200 nm, which was chosen on the basis of the experimentally observed intersubunit distance distribution (Fig. 5F). Under these conditions, a lifetime of 8.8 s was obtained (fig. S18), which is in excellent agreement with the lifetime obtained by the basic (single compartmentalization) MSK model (Fig. 6B).
Fig. 6. Complex reassociation in hierarchical confinement zones.
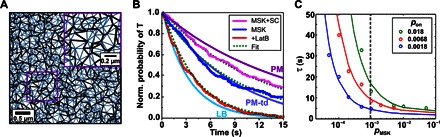
(A) Spatial organization of confinement zones used for the model. Thin and bold lines correspond to hopping probabilities pMSK and pSC, respectively. Inset is a magnification of the region highlighted by the rectangle. (B) Comparison of apparent complex stability obtained by simulations with experimentally observed lifetimes plotted as exponential decay curves [LB, molecular kinetics determined with artificial membranes in vitro; PM, complex lifetimes observed in the PM; PM-td, complex stability considering transient dissociation of dimers above a 200-nm intersubunit distance threshold chosen on the basis of the experimentally observed intersubunit distance distribution (Fig. 5F)]. (C) Role of association kinetics and hopping probability pMSK for stabilizing complexes, as measured by their lifetime (with respect to our selected metric).
Having identified a set of biologically realistic parameters describing complex reassociation, we further investigated whether cells could have developed a regulatory mechanism to exploit or control the stabilizing effect of receptor confinement. Two key parameters were taken into account: (i) the molecular two-dimensional (2D) association rate constant , which determines the reassociation probability of transiently confined interaction partners, and (ii) the MSK hop probability pMSK, which determines the confinement of receptors in the MSK. We systematically explored how MSK-induced complex stabilization depends on the binding probability along with MSK hop probabilities (Fig. 6C). As expected, an approximately threefold increase of the binding probability results in an even longer apparent complex lifetime. Upon threefold reduction of the binding probability (and, hence, ) from the experimentally determined value, stabilization by the MSK could no longer be observed at this hopping probability. A similar effect could be obtained with a fourfold increase of the hop probability, which is in line with our experimental observations upon LatB treatment.
DISCUSSION
Hierarchical organization models of the PM predict an interplay of molecular interactions and confinement into membrane microdomains. Using single-molecule localization techniques in combination with spatial stochastic modeling, we obtained evidence that MSK-based diffusion barriers play an important role in maintaining signaling complexes by facilitating reassociation of the same receptor subunits, thus ensuring sustained “signaling bursts” (18). We successfully reproduced the experimentally observed complex stabilization in a spatial stochastic model of receptor dynamics by incorporating passive diffusion barriers. All parameters used for these simulations were directly derived from experimental data, corroborating the validity of the model. Our analyses suggest that secondary confinement beyond the 70-nm MSK meshwork substantially contributed to further stabilizing the complex, which could be experimentally resolved. A similar hierarchical confinement has been recently observed for epidermal growth factor receptor dimerization (31), suggesting more general features of PM compartmentalization, the underlying molecular and cellular bases of which remain to be investigated. According to our simulations, both levels of compartmentalization are required to enhance rapid complex reassociation to a level matching the experimentally observed lifetime of individual complexes. Transient dissociation events within SCs could be resolved experimentally, thus confirming the relevance of these structures in maintaining receptor dimers. Whereas receptor stabilization by transient confinement had been previously hypothesized (18, 31), we here quantified its molecular and cellular determinants on the basis of a fully quantitative model.
Our simulations suggest that the molecular 2D association rate constant of IFNAR1 recruitment into the ternary complex is just large enough for the complex to experience a moderate increase in apparent lifetime, through repeated reassociation of its subunits. Thus, it appears that MSK permeability may have evolved to a level that supports efficient reassociation of receptor interactions with sufficiently high on-rates. Given that molecular association rate constants substantially higher than those reported for the IFNAR1-IFNα2 complex are frequently observed, even stronger MSK effects on the reassociation of receptor dimers are possible. However, for the cell, this reassociation of dynamic signaling complexes seems to establish another regulatory control mechanism because the lifetime of a functional signaling complex is defined not only by molecular interaction dynamics but also by cellular context. We found that MSK integrity is important for efficient downstream signaling by the IFN receptor, implicating a key role of receptor dimer stability in effector activation. IFNβ binds IFNAR1 with an approximately 100-fold higher binding affinity and complex stability than IFNα2, a feature that has been correlated with distinct cellular activities of IFNβ (46). These differential activities of IFNα2 and IFNβ strongly depend on the cellular context (47), which may relate to a differential degree of MSK compartmentalization (3, 6). Thus, the regulation of complex stability via the MSK permeability emerges as an important regulatory determinant of the functional plasticity of cytokine receptors and receptor tyrosine kinases, a prominent feature of signal transduction based on receptor dimerization.
MATERIALS AND METHODS
Materials
Poly(ethylene glycol) (PEG)–amino functionalized QDs (QD605 and QD655; cat. no. Q21501MP and Q21521MP, respectively) were purchased from Invitrogen and were functionalized as monovalent biotin QDs, as recently described (26). Poly-l-lysine-graft-(PEG) functionalized with an RGD peptide (PLL-PEG-RGD) (48) for minimizing nonspecific binding of QDs on the surface was synthesized, as recently detailed (40). Bovine serum albumin (BSA) (essentially fat acid–free) was purchased from Sigma-Aldrich. Halo-PEG4-biotin ligand (HTL-biotin) was purchased from Promega (cat. no. G859). Benzylguanine-biotin (SNAP-biotin) was purchased from New England Biolabs (cat. no. S9110S). Glucose oxidase and glucose were purchased from Sigma, and glucose catalase was from Roche Applied Science. LatB from Latrunculia magnifica was purchased from Sigma-Aldrich (L5288). If not specified otherwise, all other materials were purchased from Sigma-Aldrich.
Protein expression and purification
IFNα2 was expressed and purified as previously described (22). bSAV was prepared by fractionation of streptavidin tetramers that were obtained by refolding a mixture of dead and alive streptavidin monomers, as previously described (49). Expression plasmids for dead and alive streptavidin monomers were based on vectors obtained from A. Ting (Massachusetts Institute of Technology, Cambridge, MA) but included a tobacco etch virus (TEV) cleavage to allow the removal of the His tag (50). After incubation with the TEV protease, bSAV was purified by immobilized metal ion affinity chromatography.
Plasmids and cell culture
Plasmids for the expression of IFNAR1 fused to the HaloTag (51) and of IFNAR2 fused to the optimized SNAP tag (52) were generated as follows: The genes of full-length IFNAR1 and IFNAR2 without the N-terminal signal sequences were inserted into pDisplay (Invitrogen) via the Bgl II and Pst I restriction sites, respectively. Subsequently, genes coding for the HaloTag and the SNAP tag were inserted via the Bgl II site. The constructs, including the signal sequence of the pDisplay vector (immunoglobulin κ chain leader sequence), were transferred by restriction with Eco RI and Not I into modified versions of pSems-26m (Covalys Biosciences), linking the open reading frame to a neomycin or puromycin resistance cassette, respectively, via an internal ribosomal entry site. STAT2 fused to a C-terminal TagRFP (STAT2-TagRFP) was cloned into the pSEMS-26m vector (Covalys Biosciences) by cassette cloning, as recently described (53).
Cells were cultured at 37°C and 5% CO2 in Earle’s minimum essential medium (MEM) with stable glutamine (FG0325, Biochrom AG) supplemented with 10% fetal bovine serum (S0615, Biochrom AG), 1% nonessential amino acids (M11003, PAA Laboratories GmbH), and 1% Hepes buffer without the addition of antibiotics. U5A cells stably transfected with HaloTag-IFNAR1 and SNAP-IFNAR2c were generated in two steps: In a first step, U5A cells were transfected by HaloTag-IFNAR1 via G418 selection. Transfected cells were selected for stable neomycin resistance by cultivation in the presence of G418 (800 μg/ml; 345810, Calbiochem). A cell clone with homogeneous and moderate expression of HaloTag-IFNAR1 was chosen and proliferated. In a second step, SNAP-IFNAR2c was transfected and selected via Puromycin resistance at 0.4 μg/ml (D00123816, Calbiochem). After transfection with STAT2-TagRFP, HeLa cells stably expressing STAT2-TagRFP were selected on the basis of the neomycin resistance.
Quantification of STAT2 translocation and STAT1 phosphorylation
IFN-stimulated STAT2 translocation in the nucleus was monitored by confocal laser scanning microscopy (IX81, Olympus, equipped with an Olympus FluoView 1000 LSM unit) at room temperature. HeLa cells stably expressing STAT2-TagRFP were cultivated on microscope coverslips. IFNα2 (20 nM) was added in the solution after the cell sample was mounted on the microscope. Fluorescence intensity of STAT2-TagRFP per pixel for the selected region of interest (ROI) in the nucleus and cytosol was obtained as Fnucl(t) and Fcyto(t), respectively. To calculate the relative fluorescence intensity (FR), an ROI outside the cell was chosen, with its intensity used as reference (Fref): FR(t) = [Fnucl(t) − Fref]/ [Fcyto(t) − Fref]. To explore the effect of MSK to STAT2 translocation, 10 μM LatB was added for 10 min at room temperature before the addition of IFNα2.
Phosphorylation of STAT1 in the HeLa cells was analyzed by flow cytometry, as previously described (54). The HeLa cells were stimulated with 0.01, 0.1, 1, and 10 nM IFNα2. Intracellular staining of pSTAT1 was performed after the permeabilization of cells with ice-cold methanol [100% (v/v)]. Alexa Fluor 488–conjugated antibodies specific for pSTAT1 were purchased from BD Biosciences and used at a 1:50 dilution. After staining, pSTAT1 was quantified by flow cytometry using an Accuri C6 flow cytometer. The extent of STAT1 phosphorylation was calculated by subtracting the mean fluorescence intensity (MFI) of pSTAT1 of the stimulated samples from that of the unstimulated samples. To evaluate the effect of destruction of the cell MSK, the same experiments were carried out after a 10-min treatment with 10 μM LatB.
QD labeling in living cells
Before labeling, cells were transferred onto glass coverslips precoated with PLL-PEG-RGD to minimize nonspecific binding of dyes and QDs on the surface, as recently detailed (40). PEG-amino functionalized QDs (QD605 and QD655, Invitrogen) were monobiotinylated (BTQD), as previously described (26). U5A cells stably expressing HaloTag-IFNAR1 and SNAP-IFNAR2 were seeded on PLL-PEG-RGD–coated glass coverslips for 48 hours before labeling. Cell surface receptors were labeled by monobiotinylated QDs in two steps. First, the HaloTag was biotinylated by incubating 1 μM HTL-biotin for 45 min at room temperature. The cells were washed three times with phosphate-buffered saline (PBS)–BSA buffer (PBS containing 1% essentially fat acid–free BSA) and then incubated with 500 nM His tag–free bSAV for 10 min, followed by washing three times with PBS-BSA. Subsequently, 20 nM BTQD605 in PBS-BSA was added and incubated for 10 min. The cells were then washed twice with MEM containing an additional 0.1 mM biotin to block the remaining biotin-binding sites and washed two more times with MEM alone. The SNAP tag was then biotinylated by incubating 400 nM SNAP-biotin ligand for 10 min, followed by washing three times with PBS-BSA. Subsequent reactions with bSAV and BTQD655 were performed as described above. After the second QD labeling step, the cell sample was washed three times with MEM without phenol red for microscopy experiments.
Single-molecule fluorescence microscopy
Single-molecule fluorescence imaging was carried out with an inverted total internal reflection fluorescence (TIRF) microscope (Olympus IX71), equipped with a dual-line TIRF illumination condenser (Olympus) and a back-illuminated electron-multiplied (EM)–charge-coupled device (CCD) camera (iXon DU897D, Andor Technology). An image acquisition rate of 30 Hz was set for the EM-CCD camera. A 488-nm solid-state laser (200 mW Sapphire, Coherent) was coupled into the microscope through polarization-maintaining monomode fibers (kineFLEX, Point Source). Laser light was reflected by a quad-line dichroic beam splitter (Di R405/488/561/647, Semrock) and passed through an objective of high numerical aperture (NA) (150× TIRF objective UAPO; NA, 1.45; Olympus). A typical excitation power of 800 μW was measured at the objective. For simultaneous dual-color detection of QD605 and QD655, a DualView microimager (Optical Insight) was used together with a 640 DCXR dichroic beam splitter (Chroma) in combination with the band-pass filters FF01-585/40 and FF01 670/30 (Semrock), respectively. Typical time series of 2000 to 20,000 frames were recorded at 30 Hz (~1 to 10 min). To minimize photooxidation of QDs over a long period, an oxygen scavenging system containing glucose oxidase (0.5 mg ml−1), catalase (40 mg ml−1), and 5% (w/v) glucose was added. Under these conditions, no photooxidation of QDs was observed during the experimental period (~10 min of continuous illumination).
All live cell images were acquired at room temperature. Receptor dimerization was initiated by incubating 50 nM IFNα2, thus ensuring very rapid saturation of cell surface IFNAR2 by the ligand. Imaging was performed in the presence of IFNα2. For inhibition of actin polymerization, 10 μM LatB was added for 10 min at room temperature before imaging. For all dual-color single-molecule fluorescence imaging experiments, the overlay of the two channels was calibrated by imaging fluorescent microbeads (TetraSpeck microspheres 0.1 μm, T7279, Invitrogen), which were used for calculating a transformation matrix. After channel alignment based on the transformation matrix, the deviation between the channels was found to be 7.3 ± 4.0 nm (fig. S19).
For fast image acquisition, an sCMOS camera (ORCA-Flash 4.0 V2, Hamamatsu) was mounted on the same microscope setup. In combination with the 150× TIRF objective, an effective pixel size of 43 nm was obtained. Imaging was limited to areas of 2048 × 102 pixels, yielding a frame readout rate of 2000 Hz upon operating the camera in the frame transfer mode. During the fast QD imaging experiments, the typical output laser power at the objective was set to be 20 mW for detection of individual QDs in each frame. The typical imaging time was 15 s, which corresponds to 30,000 frames altogether.
Colocalization analysis and single-particle tracking
To localize and track individual QD-labeled receptors, a 2D Gaussian mask for localization of single emitters was used (55, 56). For colocalization analysis, particle coordinates from the two channels were aligned by a projective transformation using a transformation matrix. The transformation matrix was calculated on the basis of a calibration measurement with multicolor fluorescent beads (TetraSpeck microspheres 0.1 μm, Invitrogen) that are visible in both spectral channels (cp2tform of type “affine,” MATLAB, MathWorks). An average alignment precision of 7.3 ± 4.0 nm was obtained between the two channels (fig. S19). Particles colocalized within a distance of 50 nm were selected for colocomotion analyses.
Single-molecule trajectories were reconstructed using the multitarget tracing algorithm (57). Colocomotion trajectories were obtained by applying the same algorithm to the data set of colocalized molecules. The detected trajectories were evaluated with respect to their step length distribution to determine the diffusion constants. To minimize the effect of long-range perturbations and misconnections, we focused on local displacements in five steps (0.16 s). Hence, only trajectories observed for at least six consecutive frames were used.
Calculated displacements r were pooled for analysis. The cumulative probability of squared displacement p(r2) was nonlinear least squares fitted by a two-component equation (31, 58, 59) as
| (1) |
Here, D1 and D2 correspond to diffusion constants of a fast and a slow mobility component, respectively, and f is the fraction of the first component. For each fitting, we pooled ~1000 to 4000 trajectories recorded within 2000 frames (64 s). To exclude biases in our analyses of diffusion properties and colocalization, immobile particles were identified by the DBSCAN (density-based spatial clustering of applications with noise) algorithm (60), as recently described (44). Using optimized parameters, the detected localizations with very high spatiotemporal correlation (that is, immobile over very long times) were grouped and filtered out.
Time-lapse particle correlation
To increase cotracking fidelity, we used spatiotemporal particle correlation. A filtering program that employs particle correlation in a time-lapse manner was developed for selecting colocalized particles with coordinates of (xi, yi, ti), according to the following equation
| (2) |
Here, the left part is the square displacement of each particle relative to the averaged position; and denote the mean values of x and y coordinates of all colocalized particles; N is the total number of colocalized particles. D is the diffusion constant of QD-labeled receptors, which is 0.06 μm2/s (table S1); t is 20 frames (0.64 s), defined as tconf, a reliable time window for particle correlation in which colocalized QD pairs are considered to not have disappeared because of blinking. It was found that when the observation window is longer than 20 frames, the cumulative probability of observing the QD pairs is higher than 0.9 (fig. S20). By shifting the as-defined time window frame by frame, colocalization noise can be distinguished from true colocomotion events by spatial correlation because random colocalizations are not spatially correlated, and QD blinking can be overcome by temporal correlation (fig. S5B). After processing the time-lapse particle correlation, the noise-filtered, blinking-inhibited colocalizations were obtained. They were connected to colocomotion trajectories according to the highest connection possibility based on the same protocol previously published (9, 31).
Tracking and localization microscopy
Superresolution images of the QD-labeled receptors were obtained on the basis of TALM, as previously described (45). To generate superresolution images, a 2D Gaussian mask for localization of single emitters was used in each frame of an image stack (55, 56). The localization precision in two dimensions was determined as previously described by Thompson et al. (55). To render TALM images, the position of each molecule localized within the image stack was cumulatively plotted as a Gaussian blurred spot with the SD of its point spread function (PSF) set to the averaged localization precision (20 nm). Typical frame numbers are between 300 and 3000 frames (~10- to 96-s integrated imaging time). To explore the dynamics of superresolved structures, a subset of TALM images within a fixed time span (observation window) was reconstructed. The observation window was sliding along the whole TALM image stack in a time-lapse manner to generate time-lapse TALM (40). The typical observation window was 30 to 1000 frames in size (~1- to 32-s integrated imaging time).
pcTALM was carried out as recently described in detail (40). For the characterization of transient confinement zones, TALM images were generated from 800 frames with a time lapse of 256 ms. In the first step, a conventional image correlation spectroscopy (ICS) analysis (61, 62) was applied to the Gaussian-blurred TALM images. For pair correlation of the TALM images, the Cartesian coordinates of the ICS function were converted into polar coordinates to yield the pair correlation g(r), which was fitted by a two-component model (63)
| (3) |
where A is the amplitude of free diffusion, σ is the SD of the PSF of the Gaussian-blurred TALM images (σ = 20 nm), B is the amplitude of confined diffusion, and λ is the average radius of the randomly shaped confinement zones. Fitting g(r) by Eq. 3 yields λ, A, and B, as shown in fig. S13C. The confinement probability (cp) for describing the chance of the diffusive particle to be confined in the analyzed area was calculated as
| (4) |
Confinement and cluster analysis
QD-labeled receptors in TIRF images acquired with an sCMOS camera at an acquisition rate of 2000 Hz were localized and tracked as described above. MSD analysis according to the published methods (42, 64–66) was used to identify transiently confined receptor diffusion. Anomalous diffusion was described as
| (5) |
where α is the anomalous diffusion coefficient: α =1, Brownian diffusion; 0 ≤ α < 1, anomalous diffusion. α was determined by plotting MSD/t versus t and fitting of the equation
| (6) |
The effect of membrane compartmentalization on the diffusion in the short term (<32 ms) was identified by fitting a hop diffusion model (65)
| (7) |
where L, Dmicro, and DMACRO denote the diameter of compartment, the local diffusion constant, and the macroscopic diffusion constant, respectively, and c is a constant offset related to the spatial localization precision. The residence time within the compartment τcomp was calculated according to (3)
| (8) |
The time it takes for the molecule to experience the effect of the barrier of the confinement τbarrier was calculated according to (66)
| (9) |
Confinement zones were further identified by spatiotemporal cluster analysis of the fast TALM images with an average localization precision of 15 nm, which were rendered from 15,000 frames acquired in 7.5 s. Cluster analysis of the TALM images was based on the method of DBSCAN (60), which defines clusters relying solely on the principle of density reachability. For the quantification of the secondary confinement, the reported 70 nm as the average mesh diameter of the MSK in HeLa cells (3) was chosen to determine the density reachability. After points were classified into clusters, the residence time was quantified by counting the time span of the first point to the last point in each cluster. The MATLAB function kmeans was used to determine their sizes. All sizes and residence times of clusters were subjected to histogram analysis (fig. S15).
Simulations of diffusion and interaction dynamics in a compartmentalized PM
All simulations of the interaction dynamics of IFNAR1 (R1), IFNAR2:IFN (R2), and the ternary complex (T) were performed using the computer program Smoldyn (67). The parameter values for (i) the diffusion constants of all three species, (ii) the rate at which the complex T dissociates, and (iii) the association radius were directly obtained from experimental measurements (34, 43, 68). The parameter value for (iv) the probability of binding was fitted with irreversible binding dynamics obtained from the experiments and depended on the simulation time step. The latter was set to ensure nanometer resolution below the binding radius. MSK compartmentalization of the membrane was introduced as a random triangulation of the 2D plane. The (v) distribution of compartment sizes defined as the square roots of triangle areas and (vi) the median length scale were chosen to match those reported from experiments on representative cell lines (3). The diffusive jump of a molecule from one triangular compartment to another was accepted with a prespecified (vii) hop probability. In a second step, a higher-level (secondary) compartmentalization was established by dividing all triangles into separate groups. The outer boundaries of these larger compartments were assigned a distinct hop probability. All hop probabilities were fitted using experimentally obtained jump length distributions and, in the case of the model with SCs, MSK dwell times. Detailed information on the modeling and simulation can be found in section S1.
Supplementary Material
Acknowledgments
We thank G. Hikade and H. Kenneweg for technical support. Funding: C.Y., C.P.R., S.W., and J.P. were supported by grants (to J.P.) from the Deutsche Forschungsgemeinschaft (SFB 944) and by the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 223608 (IFNaction). T.T.M.-L. and A.L. were supported by funding from the Cabinet Office of Japan. T.T.M.-L. and A.L. would also like to thank the Isaac Newton Institute for Mathematical Sciences for support through EPSRC grant number EP/K032208/1. K.C.G. was funded by the Mather’s Foundation, HHMI, and NIH RO1-AI51321. Author contributions: J.P. conceived the project together with C.Y.; A.L., T.T.M.-L., C.Y., and J.P. designed the experiments. A.L. designed the mathematical model and computational framework. C.Y. and I.M. performed laboratory experiments. T.T.M.-L. and A.L. performed all computational simulations. S.W. generated the cell lines. C.P.R. programmed the image analysis software. C.Y., T.T.M.-L., K.C.G., A.L., and J.P. wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/12/e1600452/DC1
section S1. Description of the spatial stochastic models and the simulations
table S1. Measured IFN receptor diffusion constants under different conditions.
table S2. Calculated binding probability pon for different microscopic diffusion constant Dmicro.
table S3. Parameter scenarios (DR1/R2, Δt, ron, and pon) and mean lifetimes τ for fitting hop probabilities.
table S4. Experimental properties of primary and SCs.
fig. S1. Comparison of non-colocalization criteria.
fig. S2. Effect of the binding probability on the simulated receptor complex lifetime.
fig. S3. Simulation of ternary complex lifetime in the hierarchical plasma membrane model.
fig. S4. Orthogonal labeling of IFNAR1 and IFNAR2 with monovalent QD.
fig. S5. High-fidelity colocomotion analysis.
fig. S6. Role of the MSK for receptor diffusion.
fig. S7. Model of the stochastic MSK meshwork used for simulations.
fig. S8. Fitting of hop probabilities of the MSK model.
fig. S9. Fitting of association probabilities.
fig. S10. Sample simulations of ternary complex dissociation and reassociation.
fig. S11. Dependency of simulated receptor complex lifetime on MSK hop probability.
fig. S12. Effect of the reaction radius on the simulated receptor complex lifetime.
fig. S13. TALM and pcTALM analyses of receptor confinement.
fig. S14. Primary and secondary confinement confirmed by tracking receptors at 2000 Hz.
fig. S15. Compartment size and dwell time determined by DBSCAN analysis of fast TALM images.
fig. S16. Spatial stochastic model for simulating hierarchical confinement zones.
fig. S17. Fitting of hop probabilities in the refined MSK model.
fig. S18. Experimental trajectory length histograms for different colocalization cutoffs.
fig. S19. Channel alignment for colocalization analysis.
fig. S20. Determination of the observation window for time-lapse pcTALM.
movie S1. Controls confirming the labeling specificity.
movie S2. Dual-color single-molecule imaging of QD-labeled IFNAR1 and IFNAR2 in a live cell.
movie S3. Characterization of receptor diffusion before and after IFN stimulation.
movie S4. Assembly and dissociation of ternary complexes revealed by single-molecule tracking.
movie S5. Diffusion, dissociation, and reassociation of an individual ternary complex in the MSK meshwork, as obtained from spatial simulations.
movie S6. Experimentally observed receptor rebinding events.
movie S7. Transient receptor confinement at the native cell PM observed by fast TALM.
REFERENCES AND NOTES
- 1.Sako Y., Kusumi A., Compartmentalized structure of the plasma membrane for receptor movements as revealed by a nanometer-level motion analysis. J. Cell Biol. 125, 1251–1264 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simons K., Ikonen E., Functional rafts in cell membranes. Nature 387, 569–572 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Murase K., Fujiwara T., Umemura Y., Suzuki K., Iino R., Yamashita H., Saito M., Murakoshi H., Ritchie K., Kusumi A., Ultrafine membrane compartments for molecular diffusion as revealed by single molecule techniques. Biophys. J. 86, 4075–4093 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kusumi A., Ike H., Nakada C., Murase K., Fujiwara T., Single-molecule tracking of membrane molecules: Plasma membrane compartmentalization and dynamic assembly of raft-philic signaling molecules. Semin. Immunol. 17, 3–21 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Marguet D., Lenne P.-F., Rigneault H., He H.-T., Dynamics in the plasma membrane: How to combine fluidity and order. EMBO J. 25, 3446–3457 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morone N., Fujiwara T., Murase K., Kasai R. S., Ike H., Yuasa S., Usukura J., Kusumi A., Three-dimensional reconstruction of the membrane skeleton at the plasma membrane interface by electron tomography. J. Cell Biol. 174, 851–862 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kusumi A., Shirai Y. M., Koyama-Honda I., Suzuki K. G. N., Fujiwara T. K., Hierarchical organization of the plasma membrane: Investigations by single-molecule tracking vs. fluorescence correlation spectroscopy. FEBS Lett. 584, 1814–1823 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Simons K., Gerl M. J., Revitalizing membrane rafts: New tools and insights. Nat. Rev. Mol. Cell Biol. 11, 688–699 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Andrews N. L., Lidke K. A., Pfeiffer J. R., Burns A. R., Wilson B. S., Oliver J. M., Lidke D. S., Actin restricts FcεRI diffusion and facilitates antigen-induced receptor immobilization. Nat. Cell Biol. 10, 955–963 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lasserre R., Guo X.-J., Conchonaud F., Hamon Y., Hawchar O., Bernard A.-M., Soudja S. M. H., Lenne P.-F., Rigneault H., Olive D., Bismuth G., Nunès J. A., Payrastre B., Marguet D., He H.-T., Raft nanodomains contribute to Akt/PKB plasma membrane recruitment and activation. Nat. Chem. Biol. 4, 538–547 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Jaqaman K., Kuwata H., Touret N., Collins R., Trimble W. S., Danuser G., Grinstein S., Cytoskeletal control of CD36 diffusion promotes its receptor and signaling function. Cell 146, 593–606 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dehmelt L., Bastiaens P. I. H., Spatial organization of intracellular communication: Insights from imaging. Nat. Rev. Mol. Cell Biol. 11, 440–452 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Jaqaman K., Grinstein S., Regulation from within: The cytoskeleton in transmembrane signaling. Trends Cell Biol. 22, 515–526 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cambi A., Lidke D. S., Nanoscale membrane organization: Where biochemistry meets advanced microscopy. ACS Chem. Biol. 7, 139–149 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bethani I., Skånland S. S., Dikic I., Acker-Palmer A., Spatial organization of transmembrane receptor signalling. EMBO J. 29, 2677–2688 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salaita K., Nair P. M., Petit R. S., Neve R. M., Das D., Gray J. W., Groves J. T., Restriction of receptor movement alters cellular response: Physical force sensing by EphA2. Science 327, 1380–1385 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casaletto J. B., McClatchey A. I., Spatial regulation of receptor tyrosine kinases in development and cancer. Nat. Rev. Cancer 12, 387–400 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalay Z., Fujiwara T. K., Kusumi A., Confining domains lead to reaction bursts: Reaction kinetics in the plasma membrane. PLOS ONE 7, e32948 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen B., Novick D., Barak S., Rubinstein M., Ligand-induced association of the type I interferon receptor components. Mol. Cell. Biol. 15, 4208–4214 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamken P., Lata S., Gavutis M., Piehler J., Ligand-induced assembling of the type I interferon receptor on supported lipid bilayers. J. Mol. Biol. 341, 303–318 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Thomas C., Moraga I., Levin D., Krutzik P. O., Podoplelova Y., Trejo A., Lee C., Yarden G., Vleck S. E., Glenn J. S., Nolan G. P., Piehler J., Schreiber G., Garcia K. C., Structural linkage between ligand discrimination and receptor activation by type I interferons. Cell 146, 621–632 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piehler J., Schreiber G., Biophysical analysis of the interaction of human ifnar2 expressed in E. coli with IFNα2. J. Mol. Biol. 289, 57–67 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Gavutis M., Lata S., Lamken P., Müller P., Piehler J., Lateral ligand-receptor interactions on membranes probed by simultaneous fluorescence-interference detection. Biophys. J. 88, 4289–4302 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaitin D. A., Laila Roisman C., Jaks E., Gavutis M., Piehler J., Van der Heyden J., Uze G., Schreiber G., Inquiring into the differential action of interferons (IFNs): An IFN-α2 mutant with enhanced affinity to IFNAR1 is functionally similar to IFN-β. Mol. Cell. Biol. 26, 1888–1897 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaks E., Gavutis M., Uzé G., Martal J., Piehler J., Differential receptor subunit affinities of type I interferons govern differential signal activation. J. Mol. Biol. 366, 525–539 (2007). [DOI] [PubMed] [Google Scholar]
- 26.You C., Wilmes S., Richter C. P., Beutel O., Liße D., Piehler J., Electrostatically controlled quantum dot monofunctionalization for interrogating the dynamics of protein complexes in living cells. ACS Chem. Biol. 8, 320–326 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Pinaud F., Clarke S., Sittner A., Dahan M., Probing cellular events, one quantum dot at a time. Nat. Methods 7, 275–285 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Michalet X., Pinaud F. F., Bentolila L. A., Tsay J. M., Doose S., Li J. J., Sundaresan G., Wu A. M., Gambhir S. S., Weiss S., Quantum dots for live cells, in vivo imaging, and diagnostics. Science 307, 538–544 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lidke D. S., Nagy P., Heintzmann R., Arndt-Jovin D. J., Post J. N., Grecco H. E., Jares-Erijman E. A., Jovin T. M., Quantum dot ligands provide new insights into erbB/HER receptor–mediated signal transduction. Nat. Biotechnol. 22, 198–203 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Dahan M., Lévi S., Luccardini C., Rostaing P., Riveau B., Triller A., Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking. Science 302, 442–445 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Low-Nam S. T., Lidke K. A., Cutler P. J., Roovers R. C., van Bergen enHenegouwen P. M. P., Wilson B. S., Lidke D. S., ErbB1 dimerization is promoted by domain co-confinement and stabilized by ligand binding. Nat. Struct. Mol. Biol. 18, 1244–1249 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kusumi A., Tsunoyama T. A., Hirosawa K. M., Kasai R. S., Fujiwara T. K., Tracking single molecules at work in living cells. Nat. Chem. Biol. 10, 524–532 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Wilmes S., Beutel O., Li Z., Francois-Newton V., Richter C. P., Janning D., Kroll C., Hanhart P., Hötte K., You C., Uzé G., Pellegrini S., Piehler J., Receptor dimerization dynamics as regulatory valve for plasticity of type I interferon signaling. J. Cell Biol. 209, 579–593 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gavutis M., Jaks E., Lamken P., Piehler J., Determination of the 2-dimensional interaction rate constants of a cytokine receptor complex. Biophys. J. 90, 3345–3355 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lenne P.-F., Wawrezinieck L., Conchonaud F., Wurtz O., Boned A., Guo X. J., Rigneault H., He H. T., Marguet D., Dynamic molecular confinement in the plasma membrane by microdomains and the cytoskeleton meshwork. EMBO J. 25, 3245–3256 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stark G. R., Kerr I. M., Williams B. R. G., Silverman R. H., Schreiber R. D., How cells respond to interferons. Annu. Rev. Biochem. 67, 227–264 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Brierley M. M., Fish E. N., Stats: Multifaceted regulators of transcription. J. Interferon Cytokine Res. 25, 733–744 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Au-Yeung N., Mandhana R., Horvath C. M., Transcriptional regulation by STAT1 and STAT2 in the interferon JAK-STAT pathway. JAKSTAT 2, e23931 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kessler D. S., Veals S. A., Fu X. Y., Levy D. E., Interferon-alpha regulates nuclear translocation and DNA-binding affinity of ISGF3, a multimeric transcriptional activator. Genes Dev. 4, 1753–1765 (1990). [DOI] [PubMed] [Google Scholar]
- 40.You C., Richter C. P., Löchte S., Wilmes S., Piehler J., Dynamic submicroscopic signaling zones revealed by pair correlation tracking and localization microscopy. Anal. Chem. 86, 8593–8602 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Droescher M., Begitt A., Marg A., Zacharias M., Vinkemeier U., Cytokine-induced paracrystals prolong the activity of signal transducers and activators of transcription (STAT) and provide a model for the regulation of protein solubility by small ubiquitin-like modifier (SUMO). J. Biol. Chem. 286, 18731–18746 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujiwara T., Ritchie K., Murakoshi H., Jacobson K., Kusumi A., Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell Biol. 157, 1071–1081 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roder F., Wilmes S., Richter C. P., Piehler J., Rapid transfer of transmembrane proteins for single molecule dimerization assays in polymer-supported membranes. ACS Chem. Biol. 9, 2479 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Waichman S., Roder F., Richter C. P., Birkholz O., Piehler J., Diffusion and interaction dynamics of individual membrane protein complexes confined in micropatterned polymer-supported membranes. Small 9, 570–577 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Appelhans T., Richter C. P., Wilkens V., Hess S. T., Piehler J., Busch K. B., Nanoscale organization of mitochondrial microcompartments revealed by combining tracking and localization microscopy. Nano Lett. 12, 610–616 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Piehler J., Thomas C., Garcia K. C., Schreiber G., Structural and dynamic determinants of type I interferon receptor assembly and their functional interpretation. Immunol. Rev. 250, 317–334 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brierley M. M., Fish E. N., Review: IFN-α/β receptor interactions to biologic outcomes: Understanding the circuitry. J. Interferon Cytokine Res. 22, 835–845 (2002). [DOI] [PubMed] [Google Scholar]
- 48.VandeVondele S., Vörös J., Hubbell J. A., RGD-grafted poly-L-lysine-graft-(polyethylene glycol) copolymers block non-specific protein adsorption while promoting cell adhesion. Biotechnol. Bioeng. 82, 784–790 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Chen I., Howarth M., Lin W., Ting A. Y., Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat. Methods 2, 99–104 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Clarke S., Pinaud F., Beutel O., You C., Piehler J., Dahan M., Covalent monofunctionalization of peptide-coated quantum dots for single-molecule assays. Nano Lett. 10, 2147–2154 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Los G. V., Encell L. P., McDougall M. G., Hartzell D. D., Karassina N., Zimprich C., Wood M. G., Learish R., Ohana R. F., Urh M., Simpson D., Mendez J., Zimmerman K., Otto P., Vidugiris G., Zhu J., Darzins A., Klaubert D. H., Bulleit R. F., Wood K. V., HaloTag: A novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 3, 373–382 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Gautier A., Juillerat A., Heinis C., Corrêa I. R. Jr, Kindermann M., Beaufils F., Johnsson K., An engineered protein tag for multiprotein labeling in living cells. Chem. Biol. 15, 128–136 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Löchte S., Waichman S., Beutel O., You C., Piehler J., Live cell micropatterning reveals the dynamics of signaling complexes at the plasma membrane. J. Cell Biol. 207, 407–418 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moraga I., Richter D., Wilmes S., Winkelmann H., Jude K., Thomas C., Suhoski M. M., Engleman E. G., Piehler J., Garcia K. C., Instructive roles for cytokine-receptor binding parameters in determining signaling and functional potency. Sci. Signal. 8, ra114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson R. E., Larson D. R., Webb W. W., Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 82, 2775–2783 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gould T. J., Verkhusha V. V., Hess S. T., Imaging biological structures with fluorescence photoactivation localization microscopy. Nat. Protoc. 4, 291 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sergé A., Bertaux N., Rigneault H., Marguet D., Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nat. Methods 5, 687–694 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Schütz G. J., Schindler H., Schmidt T., Single-molecule microscopy on model membranes reveals anomalous diffusion. Biophys. J. 73, 1073–1080 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Keijzer S., Sergé A., van Hemert F., Lommerse P. H. M., Lamers G. E. M., Spaink H. P., Schmidt T., Snaar-Jagalska B. E., A spatially restricted increase in receptor mobility is involved in directional sensing during Dictyostelium discoideum chemotaxis. J. Cell Sci. 121, 1750–1757 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Sander J., Ester M., Kriegel H.-P., Xu X., Density-based clustering in spatial databases: The algorithm GDBSCAN and its applications. Data Min. Knowl. Disc. 2, 169–194 (1998). [Google Scholar]
- 61.Petersen N. O., Höddelius P. L., Wiseman P. W., Seger O., Magnusson K. E., Quantitation of membrane receptor distributions by image correlation spectroscopy: Concept and application. Biophys. J. 65, 1135–1146 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kolin D. L., Wiseman P. W., Advances in image correlation spectroscopy: Measuring number densities, aggregation states, and dynamics of fluorescently labeled macromolecules in cells. Cell Biochem. Biophys. 49, 141–164 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Veatch S. L., Machta B. B., Shelby S. A., Chiang E. N., Holowka D. A., Baird B. A., Correlation functions quantify super-resolution images and estimate apparent clustering due to over-counting. PLOS ONE 7, e31457 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Daumas F., Destainville N., Millot C., Lopez A., Dean D., Salomé L., Confined diffusion without fences of a G-protein-coupled receptor as revealed by single particle tracking. Biophys. J. 84, 356–366 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meilhac N., Le Guyader L., Salomé L., Destainville N., Detection of confinement and jumps in single-molecule membrane trajectories. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 73, 011915 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Clausen M. P., Lagerholm B. C., Visualization of plasma membrane compartmentalization by high-speed quantum dot tracking. Nano Lett. 13, 2332–2337 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Andrews S. S., Bray D., Stochastic simulation of chemical reactions with spatial resolution and single molecule detail. Phys. Biol. 1, 137–151 (2004). [DOI] [PubMed] [Google Scholar]
- 68.Li Z., Strunk J. J., Lamken P., Piehler J., Walz T., The EM structure of a type I interferon–receptor complex reveals a novel mechanism for cytokine signaling. J. Mol. Biol. 377, 715–724 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saffman P. G., Delbrück M., Brownian motion in biological membranes. Proc. Natl. Acad. Sci. U.S.A. 72, 3111–3113 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rakickas T., Gavutis M., Reichel A., Piehler J., Liedberg B., Valiokas R., Protein-protein interactions in reversibly assembled nanopatterns. Nano Lett. 8, 3369–3375 (2008). [DOI] [PubMed] [Google Scholar]
- 71.Kusumi A., Fujiwara T. K., Morone N., Yoshida K. J., Chadda R., Xie M., Kasai R. S., Suzuki K. G. N., Membrane mechanisms for signal transduction: The coupling of the meso-scale raft domains to membrane-skeleton-induced compartments and dynamic protein complexes. Semin. Cell Dev. Biol. 23, 126–144 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/12/e1600452/DC1
section S1. Description of the spatial stochastic models and the simulations
table S1. Measured IFN receptor diffusion constants under different conditions.
table S2. Calculated binding probability pon for different microscopic diffusion constant Dmicro.
table S3. Parameter scenarios (DR1/R2, Δt, ron, and pon) and mean lifetimes τ for fitting hop probabilities.
table S4. Experimental properties of primary and SCs.
fig. S1. Comparison of non-colocalization criteria.
fig. S2. Effect of the binding probability on the simulated receptor complex lifetime.
fig. S3. Simulation of ternary complex lifetime in the hierarchical plasma membrane model.
fig. S4. Orthogonal labeling of IFNAR1 and IFNAR2 with monovalent QD.
fig. S5. High-fidelity colocomotion analysis.
fig. S6. Role of the MSK for receptor diffusion.
fig. S7. Model of the stochastic MSK meshwork used for simulations.
fig. S8. Fitting of hop probabilities of the MSK model.
fig. S9. Fitting of association probabilities.
fig. S10. Sample simulations of ternary complex dissociation and reassociation.
fig. S11. Dependency of simulated receptor complex lifetime on MSK hop probability.
fig. S12. Effect of the reaction radius on the simulated receptor complex lifetime.
fig. S13. TALM and pcTALM analyses of receptor confinement.
fig. S14. Primary and secondary confinement confirmed by tracking receptors at 2000 Hz.
fig. S15. Compartment size and dwell time determined by DBSCAN analysis of fast TALM images.
fig. S16. Spatial stochastic model for simulating hierarchical confinement zones.
fig. S17. Fitting of hop probabilities in the refined MSK model.
fig. S18. Experimental trajectory length histograms for different colocalization cutoffs.
fig. S19. Channel alignment for colocalization analysis.
fig. S20. Determination of the observation window for time-lapse pcTALM.
movie S1. Controls confirming the labeling specificity.
movie S2. Dual-color single-molecule imaging of QD-labeled IFNAR1 and IFNAR2 in a live cell.
movie S3. Characterization of receptor diffusion before and after IFN stimulation.
movie S4. Assembly and dissociation of ternary complexes revealed by single-molecule tracking.
movie S5. Diffusion, dissociation, and reassociation of an individual ternary complex in the MSK meshwork, as obtained from spatial simulations.
movie S6. Experimentally observed receptor rebinding events.
movie S7. Transient receptor confinement at the native cell PM observed by fast TALM.


