Fig. 2.
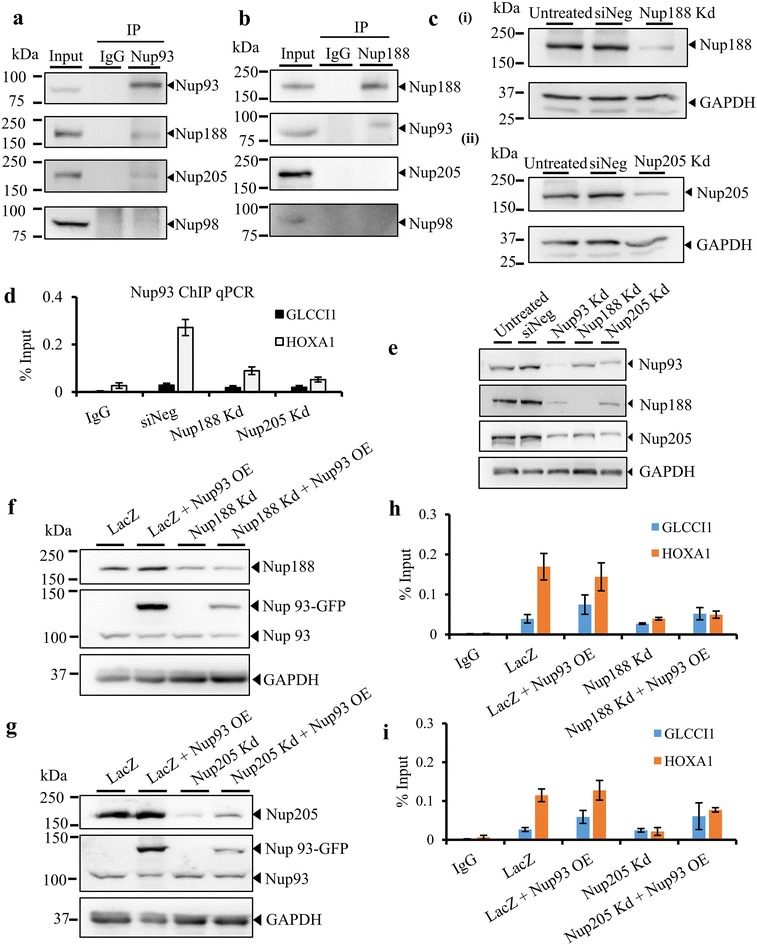
Nup93 interacts with Nup188 and Nup205 and associates with the HOXA1 promoter. a, b Immunoprecipitation was performed using antibodies specific for (a) Nup93; (b) Nup188 and IgG followed by Western blotting for Nup93, Nup188, Nup205 and negative control—Nup98 (representative data from three independent biological replicates, N = 3, single experiment for Nup98). c (i) Nup188 and (ii) Nup205 were knocked down in DLD1 cells using siRNA. A representative Western blot showing the extent of knockdown (representative Western blot from three independent biological replicates, N = 3). d ChIP experiment was performed using an anti-Nup93 antibody in untreated, non-targeting siRNA control (siNeg), Nup188 Kd (Knockdown) and Nup205 Kd cells. ChIP-qPCR analysis was used to determine the extent of Nup93 association with the HOXA1 promoter in Nup188 and Nup205 knockdown cells (Input and PanH3 in Fig. 2d are from Nup205 Kd sample) Y-axis: immunoprecipitated DNA relative to 1% input, corrected for ChIP using non-specific IgG (N = 2, data from two independent biological replicates that include a total of six technical replicates), error bar: standard error of mean (SEM). e A representative Western blot showing the effect of Nup93, Nup188 and Nup205 depletion on one another (three independent biological replicates, N = 3), f, g a representative Western blot showing overexpression of Nup93 upon Nup188 (f) and Nup205 knockdown (g). GAPDH was used as a loading control. h, i ChIP-qPCR was performed upon overexpression of (f) Nup93 in Nup188- and (g) Nup205-depleted cells. Y-axis: immunoprecipitated DNA relative to 1% input, corrected for ChIP using non-specific IgG (N = 2, data from two independent biological replicates that include a total of six technical replicates), error bar: standard error of mean (SEM)
