Abstract
Many drugs targeting the DNA damage response are being developed as anti-cancer therapies, either as single agents or in combination with ionising radiation or other cytotoxic agents. Numerous clinical trials in this area are either in progress or planned. However, concerns remain about the potential of such treatments to increase toxicity to normal tissues. In order to address this issue, we have created a novel reporter mouse line through the simultaneous incorporation of multiple reporters, β-galactosidase and firefly luciferase, into the DNA damage-inducible p21 locus. We show that in situ β-galactosidase staining facilitates high fidelity mapping of p21 expression across multiple organs and tissues at single-cell resolution, whereas the luciferase reporter permits non-invasive bioluminescent imaging of p21 expression. Using this model, we have studied the capacity of a number of DNA damaging agents, including ionizing radiation, cisplatin, and etoposide to induce p21 expression in normal tissues. We have also studied the PARP inhibitor olaparib alone or in combination with ionizing radiation as well as cisplatin. A single exposure to olaparib alone caused DNA damage to cells in the mucosal layer lining mouse large intestine. It also exacerbated DNA damage induced in this organ and the kidney by co-administered ionizing radiation. These studies suggest that olaparib might be a carcinogen in man and illustrate the power of our new model to evaluate the safety of new therapeutic regimens involving combination therapies.
Keywords: DNA damage, CDKN1A, olaparib, PARP inhibitors, anticancer drugs
Introduction
Genomic instability is a defining characteristic which distinguishes tumours from normal cells(1). For this reason, inhibitors of DNA damage response (DDR) pathways have become an important emerging class of anticancer drugs. Work in this area was given added impetus by the discovery that inhibitors of PARP1, a protein involved in single-strand DNA repair(2), are over 1000-fold more toxic to BRCA-deficient cells than normal cells in culture (3,4). This synthetic lethality suggested that PARP inhibitors, and perhaps other DDR-targeted agents, would make safe and efficacious anti-cancer drugs. One such inhibitor, olaparib(5), has been shown to be well-tolerated in clinical trials and exhibits favourable activity against ovarian(6) tumours bearing defective BRCA alleles. On this basis, it was approved in 2014 for the monotherapy of this disease (7).
Olaparib is now in clinical trials in combination with DNA damaging agents such as ionizing radiation (IR), cisplatin, gemcitabine, temozolomide and irinotecan (https://clinicaltrials.gov/ct2/results?term=OLAPARIB&pg=1) in the hope that it will synergize, and enhance the efficacy of these DNA-damaging agents selectively in tumour cells (8). In addition, on the basis that the safety of olaparib has already been firmly established, it has been proposed as a chemopreventive agent for use in healthy individuals carrying germline BRCA mutations who are at high-risk of developing ovarian and/or breast cancer (9), It has also been proposed as an agent to alleviate the symptoms of chronic inflammation (10).
However, the safety of olaparib and other DDR-targeted drugs has not yet been adequately assessed in vivo. The major concern is that they might accelerate the background rate of DNA damage in normal tissues (1). Unfortunately, the size of this risk remains unknown at present because sophisticated pre-clinical safety models with which to measure DNA damage in vivo have not been available (1).
To fulfil this medical need, we created a new and improved p21 reporter mouse line. The cell-cycle inhibitor protein p21, the product of the Cyclin-Dependent Kinase Inhibitor 1A (CDKN1A) gene, accumulates in vivo in response to many forms of DNA damage(11,12) and, as such, is an excellent in vivo biomarker. However, previous p21 reporter lines have limitations that make them unsuitable for this purpose. They either do not track p21 expression with sufficient fidelity(13) or they do not allow the resolution of expression down to individual tissues and cells (14). Our model overcomes these problems and allows us to monitor p21 expression, and DNA damage, with a high level of fidelity and resolution. Using this model, we found that a single exposure to a clinically-relevant dose of olaparib was sufficient to cause DNA damage in the large intestine of mice. We also show that olaparib exacerbates the damaging effects of IR in the large intestine and kidney. These data demonstrate that olaparib is genotoxic in mice with important possible implications for its clinical use in both disease prevention and cancer treatment.
Materials and Methods
Detailed descriptions of chemicals and y-irradiation treatments, olaparib pharmacokinetics, immunoblots, relative quantitation of mRNA species, and clinical chemical analyses can be found at SI Materials and Methods.
p21 reporter mice
For the generation of p21 reporter mice, a T2A-LacZ-loxP-T2A-Fluc-loxP cassette was inserted between the penultimate and STOP codons in exon 3 of p21. The LoxP sites were inserted in case it proved necessary to remove the luciferase reporter sequence. The positive selection marker (puromycin resistance (PuroR)) was flanked by FRT sites to allow for removal after the successful generation of transgenic mice. The targeting vector was generated using BAC clones from the C57BL/6J RPCIB-731 BAC library and electroporated into TaconicArtemis C57BL/6NTac ES cell line Art B6/3.6. Positive clones were verified by PCR and Southern blot before being injected into blastocysts from superovulated BALB/c mice. Blastocysts were injected into pseudopregnant NMRI females, and the chimerism of offspring was evaluated by coat color. Highly chimeric mice were bred with C57BL/6 females mutant for the gene encoding Flp recombinase (C57BL/6-Tg(CAG-Flpe)2 Arte). Germline transmission was identified by the presence of black C57BL/6 offspring (G1). The gene encoding Flp was removed by further breeding to Flp− partners after successful verification of PuroR removal. All animal work was carried out in accordance with the Animal Scientific Procedures Act (1986) and after local ethical review. All mice were kept under standard animal house conditions, with free access to food and water, and 12h light/12h dark cycle. Data in this paper was obtained using female mice of between 14 and 38 weeks of age. In any given experiment, mice were age-matched to within one month of each other.
Mouse genotyping
Ear biopsies of mice 4–8 weeks old were incubated at 50 °C for 4–5 h in lysis buffer containing 75 mM NaCl, 25 mM EDTA, 1% (w/v) SDS and 100 μg/ml (39 U/mg) proteinase K (Sigma). The concentration of NaCl in the reaction was raised to 0.6 M and a chloroform extraction was performed. Two volumes of isopropyl alcohol were added to the extracted supernatant to precipitate genomic DNA (gDNA). 40 μl TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0) was added to the pellet, and subsequently gDNA was dissolved overnight at 37 °C. The typical PCR sample consisted of a 25-μl volume containing 10 pmol of the primers (4155_54_for 5'-GCTACTTGTGCTGTTTGCACC-3'; 4155_55_rev 5'-TCAAGGCTTTAGGTTCAAGTACC-3'). Each reaction also contained 1.25 U Taq DNA polymerase (Thermo-Scientific) with 10 mM dNTPs, buffer and 25 mM MgCl2. The following PCR conditions were applied: 5 min, 95 °C initial denaturation; 30 s, 95 °C cyclic denaturation; 30 s, 60 °C cyclic annealing; 1 min, 72 °C cyclic elongation for a total of 35 cycles, followed by a 10-min 72 °C elongation step. All PCR protocols were developed by TaconicArtemis. PCR amplification products were analyzed by agarose gel electrophoresis.
In vivo luciferase imaging
Imaging was performed at baseline and after chemical-dosing and/or irradiation. Reporter mice were injected ip with 5 µl/g body weight RediJect d-Luciferin (30 mg/ml, Caliper) and anaesthetized by isofluorane before being transferred into the IVIS Lumina II imaging chamber (Caliper) for bioluminescent imaging. Luminescent images (5 sec, f/stop 1.2, binning 4) and gray-scale images (2 sec, f/stop 16, binning 2) were acquired. Photon fluxes in Regions Of Interest (ROI) were quantified using the LivingImage (R) Software, version 4.3.1 (Caliper). ROI were defined as an oval from just below the forepaws to just above the genitals of each mice. Photon fluxes are expressed as photons/sec/cm2/sr. Luminescent images were rendered using the fire LUT in ImageJ, and superimposed on gray-scale photographs.
Tissue harvesting and processing for cryo-sectioning
Mice were euthanized by exposure to rising concentrations of CO2. The median lobe of the liver was fixed in 10% Neutral Buffered Formalin. The proximal 2-4 cm of duodena were fixed in 4% (w/v) paraformaldehyde. All other organs, including sagittally-cut kidney, the stomach, the proximal 2 cm of the large intestine, the thymus, the spleen, the left lung lobe, the heart and the brain were fixed in Mirsky’s fixative (National Diagnostics). Tissues were stored at 4°C, formalin-fixed ones for 4 hours, Mirsky-fixed ones overnight, before being transferred into 30% (w/v) sucrose for 24 hours. Embedding was carried out in Shandon M-1 Embedding Matrix (Thermo Scientific) in a dry ice-isopentane bath. Sectioning was performed on an OFT5000 cryostat (Bright Instrument Co). With the exception of lung and brain sections, all sections were cut at 10 μm thickness with a chamber temperature of -20°C. Lung sections were cut at 12 μm thickness with a chamber temperature of -23°C. Brain sections were cut at 20 μm thickness with a chamber temperature of -23°C.
In situ β-gal staining
Sections were thawed at room temperature and rehydrated in PBS supplemented with 2 mM MgCl2 for 5 minutes before being incubated overnight at 37°C in X-gal staining solution (PBS (pH 7.4) containing 2 mM MgCl2, 0.01% (w/v) sodium deoxycholate, 0.02% (v/v) Igepal CA630, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide and 1 mg/ml 5-bromo-4-chloro-3-indolyl β-D-galactopyranosidise). On the following day, slides were washed in PBS, counterstained in Nuclear Fast Red (Vector Laboratories) for 5 minutes, washed twice in distilled water and dehydrated through 70% and 95% ethanol before being incubated in Histoclear (VWR) for 3 minutes, air-dried and mounted in DPX mountant (Sigma). Sections were evaluated by an expert, independent clinical pathologist (Dr Shaun Walsh, Department of Pathology, Ninewells Hospital, Dundee). Dr Walsh was blinded in the case of sections from olaparib-treated mice (Fig 5). Brain sections were additionally examined by a neuroscientist (Dr John Sharkey, University of Dundee). Images were captured using a Zeiss 12 megapixel digital camera attached to a Zeiss Axio Scope.A1 microscope, and controlled by means of the AxioVision v4.5 software (Carl Zeiss). Images were rendered in ImageJ.
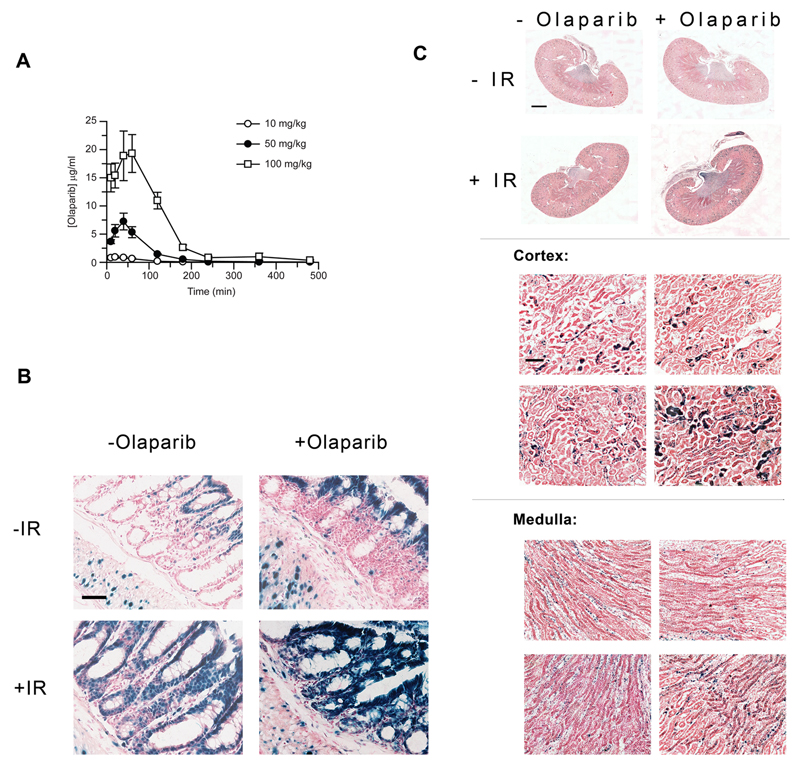
Figure 5. Effect of olaparib, alone and combined with IR, on p21 reporter activity.
A) Heterozygous reporter mice were dosed with 50 mg/kg or 100 mg/kg olaparib and drug concentrations in plasma determined at various time-points thereafter (x ± SD, n = 3 mice). B & C) p21 reporter mice were treated in triplicate with 75 mg/kg olaparib or with vehicle 30 min prior to exposure to 0- or 1 Gy IR. Representative images of in situ β-gal activity in large intestines (B) and kidneys (C). Scale bar = 50 μm for large intestine micrographs. For kidney panoramic micrographs, scale bar = 1000 μm and 100 μm for all other kidney images.
Tissue harvesting and processing for γ-H2AX staining
Mice were euthanized by exposure to rising concentrations of CO2. Organs were fixed in Gurr buffer overnight before transfer into 70% (v/v) ethanol. The following day, organs were dehydrated and embedded in paraffin and subsequently sectioned at 5 μm thickness using a Shandon Finesse 325 microtome.
γ-H2AX staining
Sections were deparaffinised in xylene and rehydrated through a graded series of 100 – 50% alcohol solutions, followed by immersion in MilliQ water. Antigen retrieval was performed using 0.01 M citrate buffer, pH 6.0. Sections were blocked in a 4% (w/v) solution of BSA in 1x TBS-T (0.01 M Tris, 150 mM NaCl, and 0.1% (v/v) Tween-20) and then incubated with 1:250 dilution of rabbit anti-γH2AX (Cell Signalling) in blocking buffer overnight at 4°C. After rinsing in 1x TBS-T, primary antibodies were detected with a 10 μg/ml solution of Alexa Fluor 488-conjugated goat anti-rabbit IgG in blocking buffer for 2 h. This solution also contained a 1:1000 dilution of the DNA counterstain DRAQ5 (Abcam). Finally, sections were mounted with Mowiol mounting medium (0.1 M Tris, pH 8.5, 10% (w/v) Mowiol 4-88, 25% (v/v) glycerol and 10 μg/ml 1,4-diazabicyclo(2,2,2)octane (DABCO)). Images were captured by confocal microscopy using the Leica S5 microscope equipped with a 40x PlanApochromat objective. Images were rendered in ImageJ.
Results
A new model for monitoring p21 expression at single-cell resolution
We generated a mouse line in which an open-reading frame encoding T2A-β-gal-T2A-luciferase was inserted between the penultimate amino acid-encoding codon and the STOP codon in exon 3 of the p21 gene (Fig 1A). T2A peptides promote a phenomenon known as ribosome skipping that allows multiple proteins to be expressed from a single mRNA(15). We therefore anticipated that three separate polypeptides, p21, β-gal, and luciferase would be expressed from this engineered allele. The β-gal protein was tagged with a nuclear localisation sequence to target it to the nucleus. Moreover, because p21 expressed from the reporter allele contains the T2A amino acid sequence at its C-terminus, it can be distinguished from p21 expressed from the wild type (wt) allele based on electrophoretic mobility.
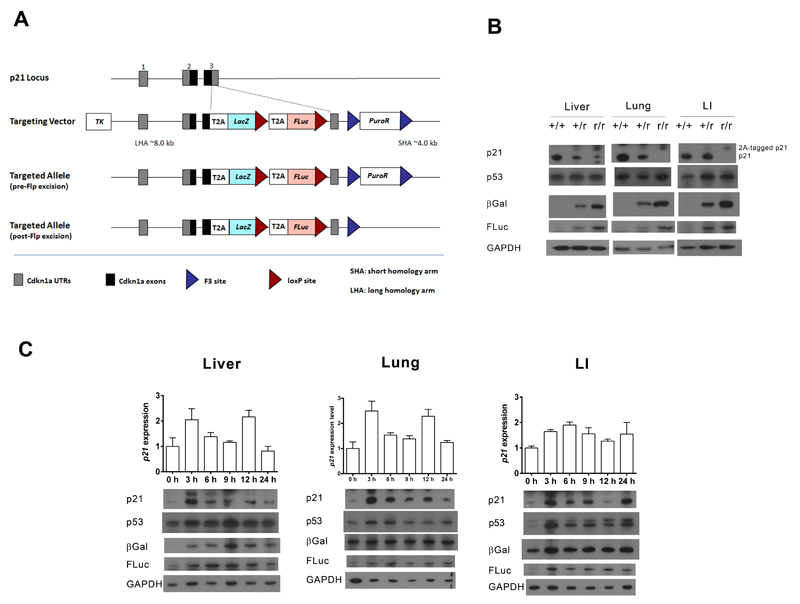
Figure 1. Validation of the p21 reporter allele.
A) The gene structures of the wild-type- and reporter p21 alleles. B) Immunoblots of pooled protein samples prepared from the liver, lung, and large intestine (LI) of groups of three wild-type (+/+), heterozygous reporter (+/r) or homozygous reporter (r/r) mice exposed to a single 4 Gy dose of IR for 6 h. Quantification of these blots can be found in Fig S1. C) Heterozygous reporter mice were exposed to a single-dose of 4 Gy of IR. At the indicated time-points post-treatment, protein and RNA were prepared from livers, lungs and large intestines (LI). The graphs depict relative p21 mRNA levels (x ± SD, n = 3 mice). Also shown are immunoblots of pooled protein samples from each group of three mice. Quantification of blot data can be found in Fig S2.
Reporter mice were fertile and produced offspring at the expected Mendelian frequencies. To establish whether three separate peptides were being correctly processed and expressed from the reporter transcript, liver, lung and large intestine lysates from irradiated mice were immunoblotted for p21, β-gal and luciferase proteins (Fig 1B). Samples of all three possible genotypes were run to confirm antibody specificity. Two bands of the predicted molecular weights for the p21 expressed from the wt allele and the T2A-tagged p21 expressed from the reporter allele were observed by western blotting (Fig 1B). Surprisingly, the level of p21 expression off the reporter allele was significantly lower than off the wt allele. Both β-gal and luciferase were detected as single proteins with higher expression levels in animals homozygous for the reporter allele. These data demonstrated that the 2A strategy for the expression of multiple reporters off a single allele worked well. However, because of the reduced expression of p21 from the reporter allele, heterozygous mice were used for all subsequent experiments, in order to minimise any unanticipated and undesirable perturbations of the DNA damage signalling. In fact, the reduced expression of p21 in the reporter line does not seem to interfere with normal responses to DNA damage as heterozygous mice express approximately half the amount of native p21 protein found in wild-type 6 h after IR treatment (Fig 1B). Given that the heterozygous reporter allele only contains one native p21 allele rather than two, this is the result we should expect if the p21 response to IR is similar in wild-type and heterozygous mice. Similarly, equivalent levels of p53 protein are found in mice of all three genotypes 6 h after exposure to IR (Fig 1B).
B-gal is expressed in the same organs, tissues, and cell types as p21
An essential attribute of any reporter system is that the pattern of expression of the reporter proteins faithfully mimic the expression of the endogenous gene. We anticipated that this would be the case in our model, as all the coding and regulatory elements that might control p21 expression are retained in the reporter allele. To confirm this, we exposed reporter mice to 4 Gy of IR and analysed expression of p21, p53, β-gal and luciferase at different time-points post-exposure (Fig 1C). Reporter activity was induced by IR concomitantly with p21 in all tissues examined. Alongside the induction of p21, a rapid and substantial accumulation of p53 protein was observed in the large intestine – but not the liver – of irradiated mice (Fig 1C), as previously described (16). We also observed a modest accumulation of p53 in lung lysates (Fig 1C). Notably, the extent of induction of p21 protein and both reporters (Fig 1C & Fig S2) exceeded that of p21 mRNA (Fig 1C), consistent with expression of p21 being determined by post-transcriptional as well as transcriptional mechanisms (11). Increased levels of reporter protein persisted longer than p21 itself. This is explained by the short p21 half-life of approximately 90 min (11) relative to luciferase (3-4 h) (17) and β-gal (>24 h) (18).
As a more stringent test of the fidelity of the reporters we measured β-gal activity at single-cell resolution across an extensive panel of organs harvested from control or irradiated mice, and compared these data with previously reported in vivo patterns of p21 expression. In view of the extent of this analysis, only representative images of β-gal staining are shown (Fig 2, S3 – S7 and SI Appendix.) These analyses showed that reporter expression patterns were the same as the current relatively limited information on p21 expression patterns in mouse tissues (see SI Appendix). Moreover, Fig S8 displays a high-resolution image of a liver section from an irradiated mouse that demonstrates both the single-cell resolution achievable with this technology and the nuclear localization of the β-galactosidase protein.
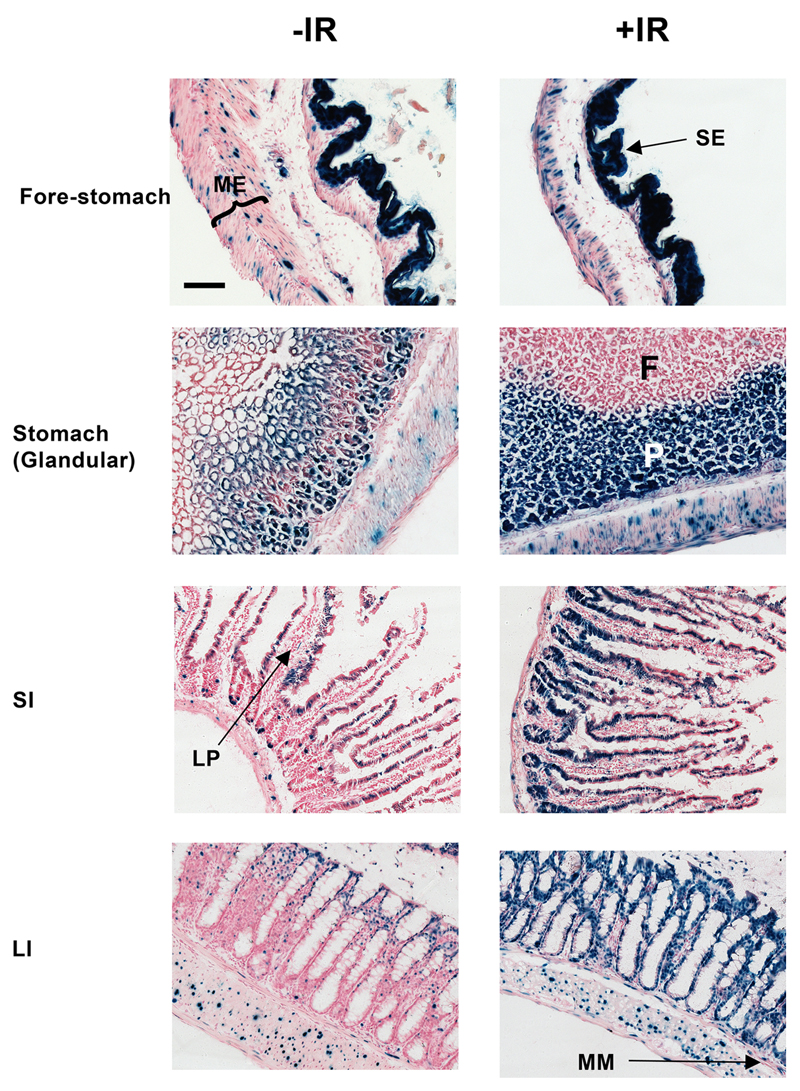
Figure 2. β-gal staining of fore-and glandular stomachs, small intestine (SI) or large intestine (LI) from untreated and irradiated p21 reporter mice.
Triplicate mice were sham-irradiated (-IR) or exposed to a single dose of 4 Gy IR (+IR) and sacrificed 24 h later. Representative images are shown. Scale-bar = 100 μm. ME, muscularis externae; SE, squamous epithelium; F, foveolar mucus-secreting cells; P, Parietal and chief cells; LP, lamina propria; MM, muscularis mucosae.
The p21 reporter is sensitive to multiple forms of DNA damage prior to overt organ toxicity
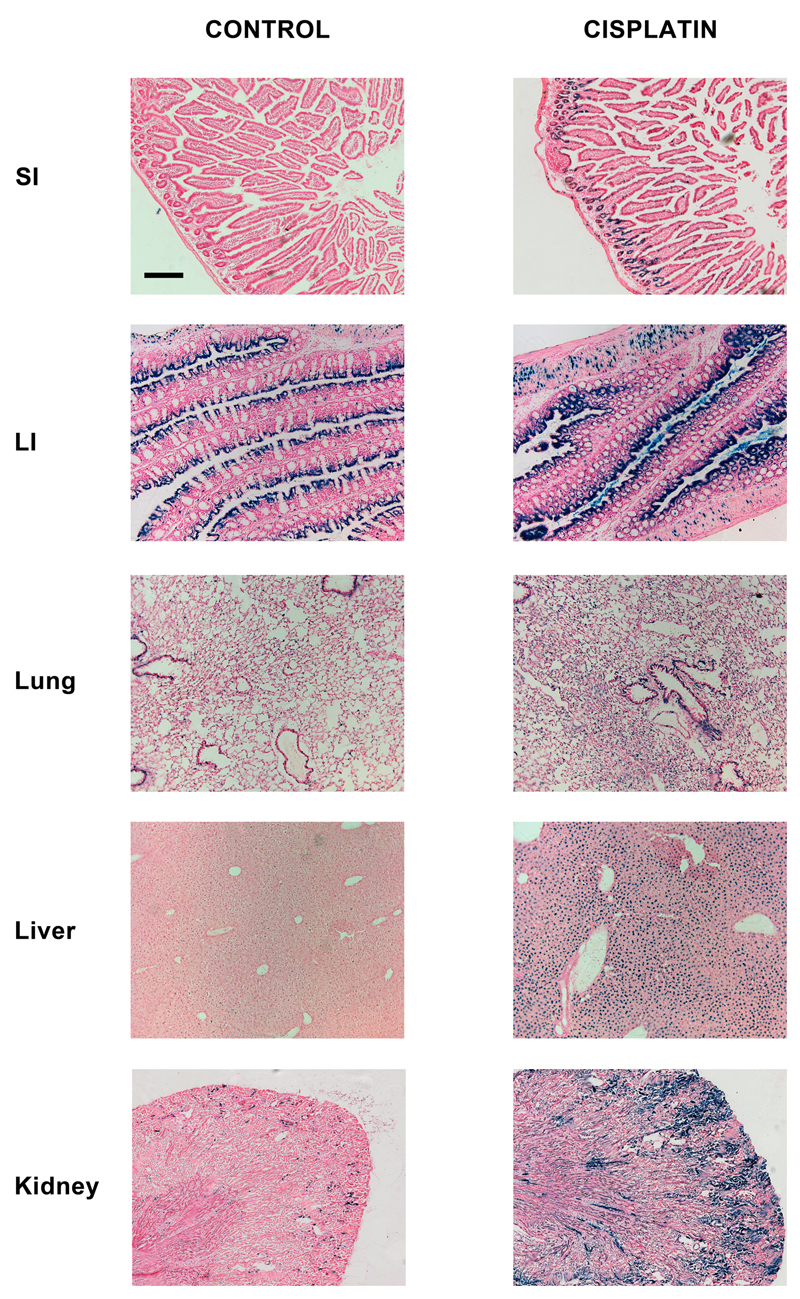
The data above demonstrate that the new mouse model is a bone fide p21 reporter and that it responds to DNA double-strand breaks caused by IR. Next, we confirmed that the model also responds to other types of DNA damage. We treated reporter mice with 0-, 1-, 3- or 10 mg/kg of cisplatin, a chemical that causes primarily DNA inter- and intra-strand crosslinks. Significant increases in the β-gal signal were observed 24 h later only at the 10 mg/kg dose of cisplatin (Fig 3 and data not shown). The pattern of induction was markedly dissimilar to that of IR. Whereas the major effect of IR, whose dose-limiting toxicity is gastro-intestinal syndrome(19), was observed in the GI tract (Fig 2), the major induction of p21 by cisplatin, which is primarily nephrotoxic(20), was observed in the kidney. Also, unlike IR, the effects of cisplatin on p21 expression in the lung were much less marked. Both agents do however elicit significant β-gal activity in hepatocytes. Importantly, the induction of reporter activity preceded overt organ damage, as clinical markers of kidney- (urea) and liver damage (alanine- and aspartate aminotransferases (Fig S9)) were not increased in these experiments.
Figure 3. β-gal staining of various organs from control and cisplatin-treated p21 reporter mice.
Mice were treated with vehicle or 10 mg/kg cisplatin and sacrificed 24 h later. Representative images are shown. Scale-bar = 200 μm.
Mice were also treated with etoposide, a topoisomerase II poison that triggers DNA double-strand breaks (21). In this case, increased reporter activity was observed predominantly in the large- and small intestines with little effect in the kidney, lung, or liver (Fig S10). This pattern of induction is consistent with clinical experience that mucositis is one of the two major dose-limiting toxicities of this drug (22). These data collectively demonstrate that these reporter mice provide an early and sensitive measure of DNA damage/toxicity in vivo.
In vivo bioluminescent detection of p21 activation
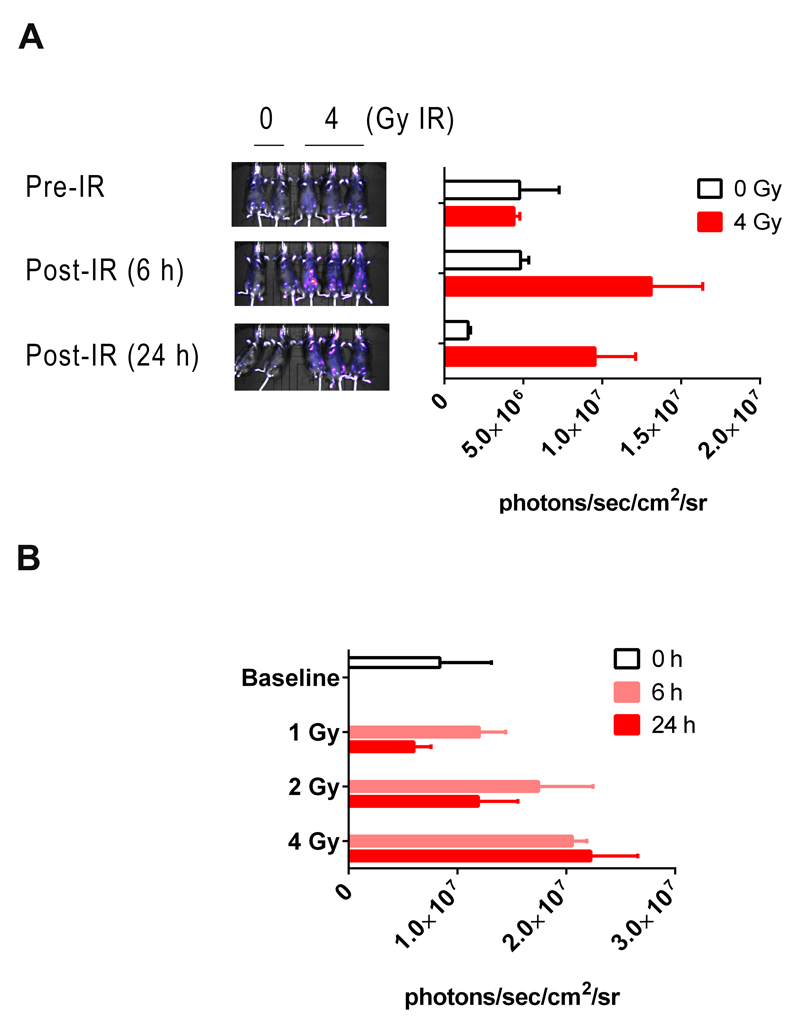
We confirmed that the luciferase reporter facilitated the non-invasive luminescent imaging of p21 induction/DNA damage. For example, we successfully measured basal as well as dose-dependent, IR-inducible luminescence in living mice (Fig 4A & B – see Fig S11 for expanded images of mice). However, the majority of the signal in both control and treated animals emanated from the skin. This was evidenced by the intense luminescence from the tail, face, paws, and genitals of the mice. In addition to these hairless ‘hotspots’, areas of high luminescence were also observed on the torso. However, these areas did not correspond to any underlying visceral tissues but were found, on closer examination, to coincide with areas where a knap of the mouse’s fur exposed the underlying skin. Although visceral organs do luminesce intensely, as evidenced by opening up the abdominal cavity (Fig S12), this cannot be observed non-invasively as these signals are attenuated by adsorption by overlying tissue and masked by the skin luminescence.
Figure 4. Bioluminescent in vivo imaging of reporter mice following IR.
A) Five heterozygous reporter mice were pre-imaged. Two of the mice were sham-irradiated whereas the remaining three were exposed to a single dose of 4 Gy of IR. Sham- (0 Gy) and irradiated mice (4 Gy) were re-imaged 6- and 24 h after exposure. Quantification of photon fluxes at the 24 h time-point is shown in the accompanying bar chart (x ± SD, n = 3 or 2 mice). B) 12 mice were in vivo imaged and randomly assigned to receive 0-, 1-, 2-, or 4 Gy of IR. Mice were in vivo imaged 6- and 24 h later (x ± SD, n = 3 mice).
Olaparib causes DNA damage alone and in combination with IR
Having validated and characterized the reporter line, we next determined whether the PARP inhibitor olaparib induces DNA damage in mouse tissues when dosed alone or in combination with IR. Pilot experiments were performed to establish the pharmacokinetic properties of olaparib so that doses were chosen to reflect the level of drug exposure in patients (Figs 5A & S13). The Cmax values for olaparib vary substantially between cancer patients, with a mean value of approximately 10 μg/ml (6). The pharmacokinetic data suggested that a dose of 75 mg/kg olaparib would be required to attain a similar level of exposure, and was used for subsequent experiments. Reporter mice were then treated with olaparib or vehicle 30 min prior to exposure to 0-, or a low dose of 1 Gy of IR. Subsequent staining for β-gal activity in liver, lungs, small intestine and brain showed that olaparib alone had no effect on p21 expression in these tissues (data not shown). In contrast, mice which had been exposed to one single dose of inhibitor had elevated levels of β-gal activity in the large intestine, primarily in the apical, terminally-differentiated epithelial cells (Fig 5B), suggesting an increased level of DNA damage in this tissue. Olaparib pre-treatment also significantly exacerbated subsequent IR-induced DNA damage in both large intestine (Fig 5B) and kidney (Fig 5C). Surprisingly, when these experiments were repeated with cisplatin (3 mg/ml) in place of IR, no potentiation of DNA damage was observed in the kidney, or in the large- and small intestines (data not shown).
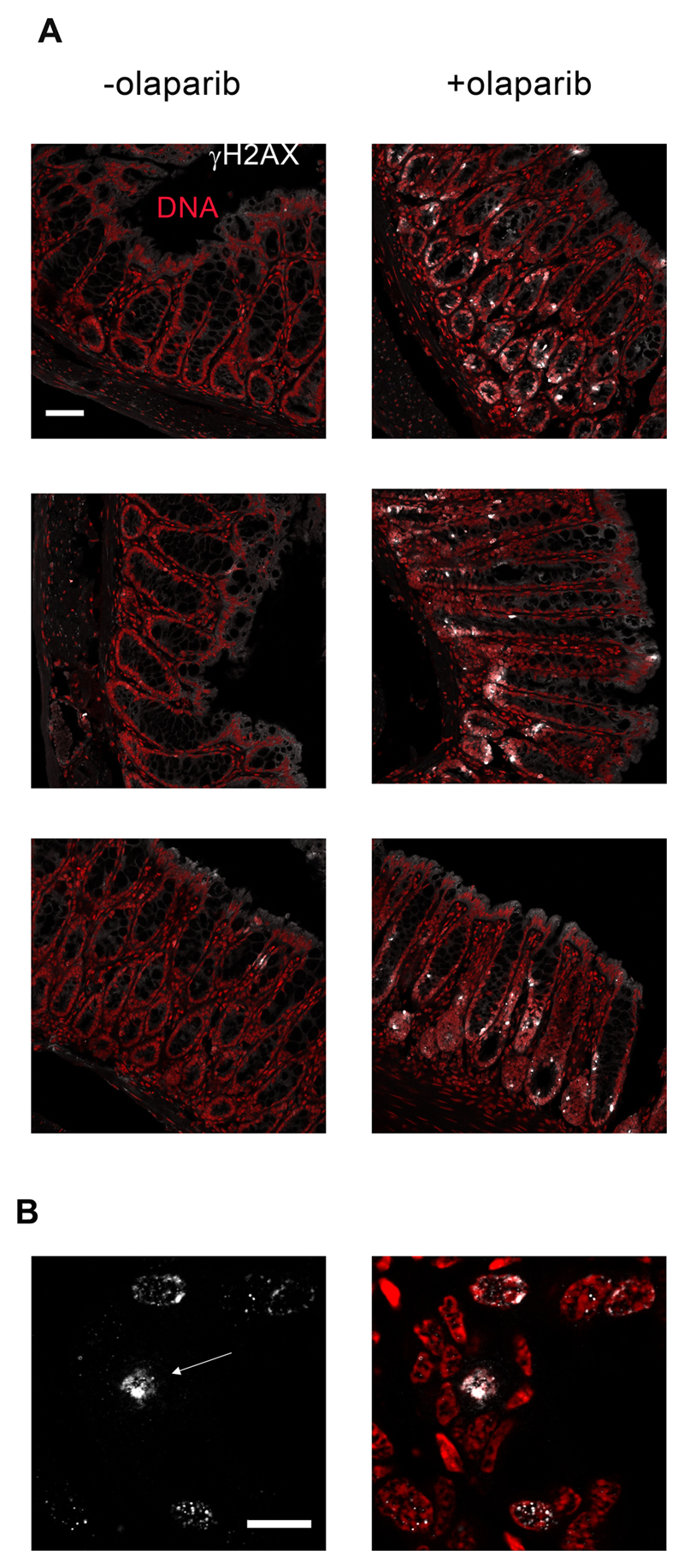
In order to confirm the reporter mouse studies that olaparib alone caused DNA damage in the large intestine, we exposed wt mice to this compound and stained large intestines for γ-H2AX, an established marker of DNA damage. Significant γ-H2AX staining in cells lining the mucosa of the large intestine were observed 4h after olaparib treatment (Fig 6A), but not at any other time point (1-, 2-, 8-, or 24 h (data not shown)). The γ-H2AX staining was largely focal indicating that it arises from DNA damage signalling (Fig 6B). We also observed a minority of cells displaying a more intense and pan-nuclear stain, the significance of which is less clear (23) but which may indicate the presence of clustered DNA lesions (24).
Figure 6. Olaparib causes DNA damage in the mucosal lining of the large intestine.
A) The micrographs display γ-H2AX staining in the large intestines of three separate untreated wild-type mice (-olaparib), or three separate mice that were euthanized 4 h after treatment with 75 mg/kg olaparib. Scale-bar = 50 μm. B) A higher resolution image demonstrating the primarily focal nature of the γ-H2AX signal in large intestines of olaparib-treated wild-type mice. The arrow points to a cell displaying a more pan-nuclear γ-H2AX signal. Scale-bar = 10 μm.
Discussion
A new p21 reporter mouse model has been developed that permits the measurement of p21 expression non-invasively in live mice (luciferase reporter) and, post-mortem, in all tissues at single-cell resolution (β-gal reporter). This has been achieved by incorporating both reporters into the p21 gene locus to create a polycistronic mRNA where each protein is translated individually by flanking each coding region with foot and mouth virus 2A sequences. It should be noted, however, that the expression of p21 from this locus was significantly reduced relative to the wild type allele. The reason for this remains unclear. One possible explanation is that translation of the polycistronic reporter mRNA is hindered by the presence of multiple T2A sequences, which cause translational pausing(15). Alternatively, it is possible that the insertion of the reporter cassette might affect mRNA secondary structure/stability (25).
We have demonstrated the versatility of the p21 reporter model by showing that it responds to both double-strand breaks (IR and etoposide) and inter- and intra-strand DNA crosslinks (cisplatin). Further work is ongoing to determine the full spectrum of DNA lesions to which it responds. The reporter is active in most organs examined (GI Tract, liver, lungs, kidneys, female reproductive tract) and in both proliferative (e.g large intestine crypt cells) and post-mitotic (e.g alveolar) cells. An advantage of our system is that, because β-gal is a stable protein, our reporter response is durable and lasts at least 24 h after damage, unlike classic markers of DNA damage, such as γ-H2AX, whose presence is often transient and easily missed.
The reporter mice were created to enable DNA damage to be measured in vivo in response to drugs and chemicals. For this first study, we investigated the effects of olaparib primarily because of the current interest in combining it with other DNA damaging agents in the treatment of cancer (1), and also because it is being considered as an agent to be used prophylactically in prevention of breast and ovarian cancer in individuals carrying BRCA mutations (26). We discovered that a single exposure to a clinically relevant dose of olaparib causes DNA damage in the large intestine of mice. Additionally, it exacerbated DNA damage induced by IR treatment in this tissue and also the kidney. Both of these findings have significant implications for the clinical use of this drug. We also note that such detrimental effects may become more pronounced, and affect other tissues, after repeat chronic dosing with olaparib.
Our finding that olaparib alone induced DNA damage in the colon suggests that the use of this drug prophylactically may increase the risk of developing colorectal cancer. This concern might seem to have been allayed by the recent report that mice chronically exposed to olaparib suffered no adverse effects (26). However, in these studies, the mice had an average lifespan of only 38 weeks due to a mammary-specific BRCA1 deletion that caused rapid breast tumour development. This lifespan may well have been too short to observe sporadic colorectal tumours, which generally present in aged mice(27). Based on our findings, the possibility that long-term exposure to olaparib is carcinogenic warrants further investigation.
At present, it is unclear whether olaparib causes DNA damage in the gut because it inhibits PARP1/2 or whether it is an off-target effect; olaparib, has broad activity and inhibits multiple PARPs in addition to PARP1/2, including PARP3/4/12/15 and 16 (28). It may also target additional proteins unrelated to the PARP family (8). However, olaparib toxicity in haploid ES cells is due to PARP1 inhibition (29). If the damage that we observed is also due to an on-target effect, it is difficult to envisage olaparib, or PARP inhibitors more generally, being developed for purposes other than the treatment of cancer.
Our finding that olaparib exacerbates IR-elicited DNA damage in normal tissues such as the large intestine and kidney, is also of significant potential clinical importance as it suggests that combination therapies involving olaparib and DNA damaging agents will increase toxicity to normal tissues. Initial clinical trial data support this contention with more adverse effects reported in patients treated with olaparib and chemotherapy than olaparib alone (30). The olaparib-induced radiosensitisation of normal tissues is particularly concerning in light of the fact that tumours are generally radiosensitised by olaparib by a factor of less than two (31). Interestingly, we observed no evidence of exacerbated DNA damage when olaparib and cisplatin were used in combination. A possible explanation is that the major DDR pathway involved in repair of cisplatin crosslinks is nucleotide excision repair (32). Unlike for the homologous recombination repair pathway involved in repairing IR-induced damage, there is no compelling evidence for a biochemical or genetic interaction between nucleotide excision repair and the PARP pathway. Therefore, the lack of an interaction between olaparib and cisplatin in our model may reflect the fact that these two drugs do not synergize together and that the rationale for combining them in the clinic is not well-founded (33). Indeed, the optimal PARP inhibitor-chemotherapy drug combination for cancer treatment remains to be determined. It will also be challenging to define the optimal doses and scheduling of combined drugs so as to minimize side-effects while maximizing efficacy. These issues could be addressed by pre-clinical studies using the p21 reporter model, especially if it were crossed onto genetically-engineered mouse models for specific cancers to permit anti-tumour efficacy to be measured alongside normal tissue toxicity.
Finally, it is worthy of note that while our motivation in developing the p21 reporter mouse was to provide a tool for pre-clinical drug safety testing, accumulation of DNA damage and p21 protein is a symptom of many degenerative diseases associated with aging, such as Hutchinson-Gifford Progeria Syndrome and Parkinson’s disease, and indeed normal aging itself(34). For this reason, monitoring of p21 levels using our reporter might catalyse research into the basic science underpinning these devastating disorders and, as an efficacy biomarker, would expedite testing of drugs to alleviate their symptoms.
Supplementary Material
Acknowledgement
We are grateful to Dr Shaun Walsh (Department of Pathology, Ninewells Hospital, Dundee) for advice in interpretation of tissue sections.
1 This work was funded by a European Research Council Advanced Investigator Award (number 294533) and a CRUK Programme grant (C4639/A10822), both awarded to CRW.
Footnotes
Disclosure of conflict of interest: The authors confirm that no potential conflict of interest exists.
References
- 1.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–94. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 2.Ashworth A, Lord CJJ, Reis-Filho JSS. Cell. Vol. 145. Elsevier Inc.; 2011. [cited 2011 Apr 3]. Genetic interactions in cancer progression and treatment; pp. 30–8. [Internet] [DOI] [PubMed] [Google Scholar]
- 3.Farmer H, McCabe N, Lord CJ, Tutt ANJ, Johnson Da, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 4.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 5.Menear Ka, Adcock C, Boulter R, Cockcroft X, Copsey L, Cranston A, et al. 4-[3-(4-cyclopropanecarbonylpiperazine-1-carbonyl)-4-fluorobenzyl]-2H-phthalazin-1-one: a novel bioavailable inhibitor of poly(ADP-ribose) polymerase-1. J Med Chem. 2008;51:6581–91. doi: 10.1021/jm8001263. [Internet] [DOI] [PubMed] [Google Scholar]
- 6.Fong PC, Boss DS, Yap Ta, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of Poly(ADP-Ribose) Polymerase in Tumors from BRCA Mutation Carriers. N Engl J Med. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 7.Deeks ED. Olaparib: First Global Approval. Drugs. 2015;75:231–40. doi: 10.1007/s40265-015-0345-6. [Internet] [DOI] [PubMed] [Google Scholar]
- 8.Ekblad T, Camaioni E, Schüler H, Macchiarulo A. PARP inhibitors: Polypharmacology versus selective inhibition. FEBS J. 2013;280:3563–75. doi: 10.1111/febs.12298. [DOI] [PubMed] [Google Scholar]
- 9.Sandhu SK, Yap Ta, de Bono JS. Eur J Cancer. Vol. 46. Elsevier Ltd; 2010. [cited 2014 Apr 16]. Poly(ADP-ribose) polymerase inhibitors in cancer treatment: a clinical perspective; pp. 9–20. [Internet] [DOI] [PubMed] [Google Scholar]
- 10.Kapoor K, Singla E, Sahu B, Naura AS. PARP inhibitor, olaparib ameliorates acute lung and kidney injury upon intratracheal administration of LPS in mice. Mol Cell Biochem. 2014;400:153–62. doi: 10.1007/s11010-014-2271-4. [Internet] [DOI] [PubMed] [Google Scholar]
- 11.Warfel Na, El-Deiry WS. p21WAF1 and tumourigenesis: 20 years after. [cited 2014 Apr 1];Curr Opin Oncol. 2013 25:52–8. doi: 10.1097/CCO.0b013e32835b639e. [Internet] [DOI] [PubMed] [Google Scholar]
- 12.Grek CL, Townsend DM, Tew KD. Pharmacol Ther. Vol. 129. Elsevier Inc.; 2011. The impact of redox and thiol status on the bone marrow: Pharmacological intervention strategies; pp. 172–84. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vasey DB, Wolf CR, MacArtney T, Brown K, Whitelaw CBa. p21-LacZ reporter mice reflect p53-dependent toxic insult. [cited 2013 Apr 12];Toxicol Appl Pharmacol. 2008 227:440–50. doi: 10.1016/j.taap.2007.11.029. [Internet] [DOI] [PubMed] [Google Scholar]
- 14.Tinkum KL, Marpegan L, White LS, Sun J, Herzog ED, Piwnica-Worms D, et al. Bioluminescence imaging captures the expression and dynamics of endogenous p21 promoter activity in living mice and intact cells. [cited 2013 Apr 12];Mol Cell Biol. 2011 31:3759–72. doi: 10.1128/MCB.05243-11. [Internet]. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3165732&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Felipe P, Luke Ga, Hughes LE, Gani D, Halpin C, Ryan MD. E unum pluribus: multiple proteins from a self-processing polyprotein. [cited 2013 Feb 28];Trends Biotechnol. 2006 24:68–75. doi: 10.1016/j.tibtech.2005.12.006. [Internet] [DOI] [PubMed] [Google Scholar]
- 16.Midgley CA, Owens B, Briscoe CV, Thomas DB, Lane DP, H P. Coupling between gamma irradiation, p53 induction and the apoptotic response depends upon cell type in vivo. J Cell Sci. 1995;108:1843–8. doi: 10.1242/jcs.108.5.1843. [DOI] [PubMed] [Google Scholar]
- 17.Leclerc G, Boockfor F, Faught W, Frawley L. Development of a destabilized firefly luciferase enzyme for measurement of gene expression. Biotechniques. 2000;29:590–1. doi: 10.2144/00293rr02. [DOI] [PubMed] [Google Scholar]
- 18.Stack J, Whitney M, Rodems S, Pollok B. A ubiquitin-based tagging system for controlled modulation of protein stability. Nat Biotechnol. 2000;18:1298–302. doi: 10.1038/82422. [DOI] [PubMed] [Google Scholar]
- 19.Kirsch DG, Santiago PM, di Tomaso E, Sullivan JM, Hou W-S, Dayton T, et al. P53 Controls Radiation-Induced Gastrointestinal Syndrome in Mice Independent of Apoptosis. Science. 2010;327:593–6. doi: 10.1126/science.1166202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhar S, Kolishetti N, Lippard SJ, Farokhzad OC. Targeted delivery of a cisplatin prodrug for safer and more effective prostate cancer therapy in vivo. Proc Natl Acad Sci U S A. 2011;108:2–7. doi: 10.1073/pnas.1011379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pommier Y, Leo E, Zhang H, Marchand C. Chem Biol. Vol. 17. Elsevier Ltd; 2010. DNA Topoisomerases and Their Poisoning by Anticancer and Antibacterial Drugs; pp. 421–33. [Internet]. Available from: http://linkinghub.elsevier.com/retrieve/pii/S1074552110001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hainsworth JD, Greco FA. Etoposide: Twenty years later. 1995 doi: 10.1093/oxfordjournals.annonc.a059180. [DOI] [PubMed] [Google Scholar]
- 23.Solier S, Pommier Y. The nuclear γ-H2AX apoptotic ring: Implications for cancers and autoimmune diseases. Cell Mol Life Sci. 2014;71:2289–97. doi: 10.1007/s00018-013-1555-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer B, Voss KO, Tobias F, Jakob B, Durante M, Taucher-Scholz G. Clustered DNA damage induces pan-nuclear H2AX phosphorylation mediated by ATM and DNA-PK. Nucleic Acids Res. 2013;41:6109–18. doi: 10.1093/nar/gkt304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pessi G, Blumer B, Haas D. lacZ fusions report gene expression, don’t they? Microbiology. 2000;147:1993–5. doi: 10.1099/00221287-147-8-1993. [DOI] [PubMed] [Google Scholar]
- 26.To C, Kim EH, Royce DB, Williams CR, Collins RM, Risingsong R, et al. The PARP inhibitors, veliparib and olaparib, are effective chemopreventive agents for delaying mammary tumor development in BRCA1-deficient mice. Cancer Prev Res. 2014;7:698–707. doi: 10.1158/1940-6207.CAPR-14-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fleet JC. Animal models of gastrointestinal and liver diseases. New mouse models for studying dietary prevention of colorectal cancer. AJP Gastrointest Liver Physiol. 2014;307:G249–59. doi: 10.1152/ajpgi.00019.2014. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wahlberg E, Karlberg T, Kouznetsova E, Markova N, Macchiarulo A, Thorsell A-G, et al. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat Biotechnol. 2012;30:283–8. doi: 10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]
- 29.Pettitt SJ, Rehman FL, Bajrami I, Brough R, Wallberg F, Kozarewa I, et al. A genetic screen using the PiggyBac transposon in haploid cells identifies Parp1 as a mediator of olaparib toxicity. [cited 2014 Apr 16];PLoS One. 2013 8:e61520. doi: 10.1371/journal.pone.0061520. [Internet]. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3636235&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oza AM, Cibula D, Benzaquen AO, Poole C, Mathijssen RHJ, Sonke GS, et al. Olaparib combined with chemotherapy for recurrent platinum-sensitive ovarian cancer: a randomised phase 2 trial. Lancet Oncol. 2015;16:87–97. doi: 10.1016/S1470-2045(14)71135-0. [Internet]. Available from: http://linkinghub.elsevier.com/retrieve/pii/S1470204514711350. [DOI] [PubMed] [Google Scholar]
- 31.Powell C, Mikropoulos C, Kaye SB, Nutting CM, Bhide SA, Newbold K, et al. Cancer Treat Rev. Vol. 36. Elsevier Ltd; 2010. Pre-clinical and clinical evaluation of PARP inhibitors as tumour-specific radiosensitisers; pp. 566–75. [Internet] [DOI] [PubMed] [Google Scholar]
- 32.Martin LP, Hamilton TC, Schilder RJ. Platinum Resistance: The Role of DNA Repair Pathways. Clin Cancer Res. 2008;14:1291–5. doi: 10.1158/1078-0432.CCR-07-2238. [Internet] [DOI] [PubMed] [Google Scholar]
- 33.Murai J, Zhang Y, Morris J, Ji J, Takeda S, Doroshow JH, et al. Rationale for poly(ADP-ribose) polymerase (PARP) inhibitors in combination therapy with camptothecins or temozolomide based on PARP trapping versus catalytic inhibition. J Pharmacol Exp Ther. 2014;349:408–16. doi: 10.1124/jpet.113.210146. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garinis GA, Van Der Horst GTJ, Vijg J, Hoeijmakers JHJ. DNA damage and ageing : new-age ideas for an age-old problem. Nat Cell Biol. 2008;10:1241–7. doi: 10.1038/ncb1108-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.