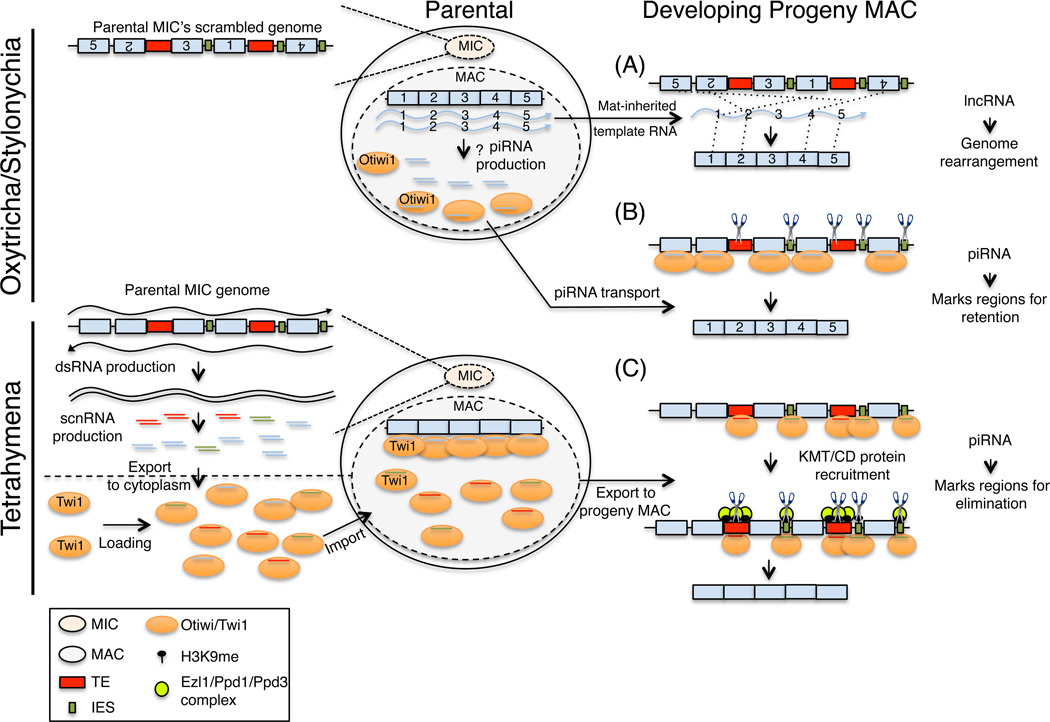
Figure 3.
ncRNA-mediated genome reorganization in ciliates. Genome rearrangement in Oxytricha involves the (A) unscrambling of the developing MAC genome. This requires the maternal inheritance of long noncoding template RNAs (blue wavy lines) from the parental MAC nucleus into the developing progeny MAC. Numbers depict segments of a gene which must be ordered to make a functional open reading frame. (B) DNA retention uses maternally inherited piRNA-Otiwi complexes (orange ovals with blue lines) to mark the regions of the genome which are retained in the developing progeny MAC nucleus. (C) DNA elimination in Tetrahymena requires the bidirectional transcription (wavy black lines) of the parental MIC genome. dsRNAs are degraded into scnRNAs (shown in red, blue and green lines) by Dcl1 and exported to the cytoplasm where they are loaded onto the PIWI protein Twi1 (orange oval). scnRNA-Twi1 complexes are imported into the parental MAC nucleus where they find the complementary genomic regions. All ‘self’ scnRNAs (blue) are eliminated, and the remaining scnRNAs (red and green), complementary to TEs (red rectangle) and IESs (green rectangle), are exported into the developing progeny MAC nucleus. scnRNA-Twi1 complexes basepair with complementary TEs and IESs in the MAC genome and recruit Ezl and Ppd1/3 proteins (lime green circle). These regions are packaged into heterochromatin and later eliminated from the MAC genome by an unknown mechanism.