Abstract
Dynamic histone modifications play an important role in controlling gene expression in response to various environmental cues. This mechanism of regulation of gene expression is important for sessile organisms, like land plants. We have previously reported consistent upregulation of various marker genes in response to gamma rays at various post-irradiation times. In the present study, we performed various chromatin modification analyses at selected loci using the standard chromatin immunoprecipitation procedure, and demonstrate that upregulation of these genes is associated with histone H3 lysine 4 tri-methylation (H3K4me3) at the gene body or transcription start sites of these loci. Further, at specific AtAgo2 loci, both H3K4me3 and histone H3 lysine 9 acetylation (H3K9ac) are important in controlling gene expression in response to gamma irradiation. There was no change in DNA methylation in these selected loci. We conclude that specific histone modification such as H3K4me3 and H3K9ac may be more important in activating gene expression in these selected loci in response to gamma irradiation than a change in DNA methylation.
Keywords: chromatin immunoprecipitation, DNA repair genes, epigenetics, gamma rays, histone modification
INTRODUCTION
Exposure of plants to ionizing radiation generates both oxidative stress and genomic stress. The most commonly used ionizing radiation in plant mutation breeding is gamma ray irradiation, which acts directly on DNA, inducing single- and double-strand breaks (DSBs). It also generates reactive oxygen species (ROS) through radiolysis of water within cells. ROS oxidize nucleotide bases, causing base substitutions and deletions. DSBs are the most lethal of the changes induced in DNA. DSBs are repaired by homologous recombination (HR) during S or G2 phase and/or by non-homologous end joining (NHEJ) during the cell cycle [1]. The repair mechanisms are regulated by ataxia–telangiectasia mutated (ATM) and ATM and Rad3-related (ATR) signalling in plants [2]. The DNA damage repair process occurs in vivo in chromatin context. Thus the role of chromatin remodelling and/or modification in the regulation of multiple signalling and downstream pathways during DNA damage repair has recently been recognized. It is well known that chromatin is modified at the N-terminals of various histone subunits (especially H3 and H4) by acetylation and methylation at lysine residues and by phosphorylation at serine/threonine residues. Histone-modifying enzymes and their specific actions have been well documented in animal cell systems and in yeast [3–5]. For example, it has been reported that a histone H2A is phosphorylated (γH2AX) as an initial response to DNA damage and targeted to DSB sites [6]. Moreover, DNA damage helps to recruit chromatin remodelling proteins through histone acetylation [7], and these proteins then activate HR or NHEJ through histone methylation [8, 9], or they recruit effector proteins involved in HR or NHEJ through histone ubiquitination in human cell lines and yeast [10, 11].
Post-translational modifications of histone N-terminal tails are known to influence the level of gene expression [12]. Enrichment of histone H3 Lys4 (H3K4me3) and acetylation of histone H3 Lys9 (H3K9ac) on the coding regions of RD29A, RD29B, RD20 and AP2 domain transcription factor occur in response to drought stress [13]. On the other hand, a decrease in methylation of histone H3 Lys27 (H3K27me3) (a negative marker for transcription) was observed at cold responsive genes COR15A and ATGOLS3 during exposure to cold temperature, along with a decrease in histone occupancy (H3-core) at promoter regions of these two genes [14]. Upregulation of various stress-inducible genes in response to high salinity, cold and abscisic acid is linked to H3 Ser10 phosphorylation, H3 Lys 14 phosphoacetylation or H4 acetylation [15]. Even the levels of H3K4me3 and H3 acetylation at coding regions of ADH1 and PDC1 increase in rice due to submergence [16]. However, there is little information on whether these histone modifications occur in plants in response to ionizing radiation.
The effect of gamma rays on gene transcription has been extensively studied in Arabidopsis, with variations observable among experiments due to differences in post-irradiation time, absorbed dose, nature of dose (chronic vs acute) and plant developmental stage and tissue type [17–19]. We previously studied gamma-ray–responsive genes at various post-irradiation times and identified several radioresponsive marker genes: At2g30360, At4g19130 and At5g24280, which showed sustainable upregulation of their transcript until 5 days after gamma irradiation [20]. Another well known gamma-ray–responsive gene, AtRad51 (At5g20850) maintained upregulated expression until 48 h [20]. At4g19130 encodes replication protein A1-E (RPA1-E), which repairs base damages specific to ionizing radiation and recruits DNA end-processing complex by acting within the ATR-dependent pathway [21]. At5g24280 encodes a gamma ray and mitomycin C–induced protein 1 (GMI1), which is involved in DSB repair through homologous recombination (HR) via the ATM pathway [22]. Similarly, AtRad51, which encodes a homologue of RecA in E. coli, is also involved in DSB repair in plants via the HR pathway. At2g30360 encodes a CBL-interacting protein kinase 11 (CIPK11), which is induced in response to gamma rays in an ATM-dependent manner and may play a role in the downstream of DNA damage response [18]. Another important player, Argonaute 2, encoded by AtAgo2 (At1g31280), is involved in DSB repair in plants and animals by recruiting DSB-induced small RNAs (diRNAs) to precisely employ other repair protein complexes at the DSB site [23]. All these important players in DNA damage repair are highly induced by gamma rays [18, 20], but the mechanism of induction in plants remains poorly understood. Although the control of gene expression mainly depends on transcriptional factors or cis-elements in promoters, eukaryotic gene expression occurs in a chromatin context, and therefore modification of histones may be necessary for the induction of gene expression in response to gamma irradiation. Among various histone modifications, we have studied active chromatin marks (H3K4me3 and H3K9ac) and repressive chromatin marks [H3K9me2, H3K27me3 and H3-core (for histone occupancy)] by chromatin immunoprecipitation (ChIP) assay, followed by ChIP-quantitative PCR (ChIP-qPCR). We report here for the first time histone modifications associated with DNA damage repair genes and correlate them with transcriptional induction of genes in response to gamma rays in Arabidopsis seedlings. We also discuss whether these dynamic chromatin modifications can be correlated with changes in DNA methylation.
MATERIALS AND METHODS
Plant material and growth conditions
Arabidopsis thaliana (Columbia-0 ecotype) seeds were surface-sterilized for 1 min in 70% ethanol and 5 min in 2% sodium hypochlorite solution, rinsed five times with sterile water, plated on one-half strength Murashige and Skoog (1/2 MS) medium with 1% sucrose and 0.6% phytoagar, and then stratified at 4°C in the dark for 3 days. Plants were grown under a 16 h light/8 h dark cycle at 23°C. Fifteen day-old seedlings were irradiated at a dose rate of 50 Gy/h for 4 h using a 60Co gamma irradiator (IR-222, MDS Nordion Inc.). The seedlings were used for experiments at 24 h after gamma irradiation in order to minimize gene induction associated with initial oxidative stress responses [20]. For genomic DNA and RNA isolation, the seedlings were frozen in liquid nitrogen and stored at −80°C. The seedlings (without root) for the ChIP experiment were harvested, washed two times with sterile distilled water and then soaked in freshly prepared 1% formaldehyde solution for 10 min in a vacuum chamber at −76 mm Hg pressure. The formaldehyde solution was neutralized with 1 M glycine for 5 min in vacuum (−76 mm Hg pressure) and washed thoroughly (two times) with sterile distilled water before storing at −80°C.
RNA isolation and real-time quantitative RT-PCR analysis
Total RNA was isolated from the seedlings stored at −80°C using the RNeasy Plant Mini Kit followed by DNase I (RNase free) treatment. The quality and quantity of isolated RNA was checked by electrophoresis on 1% agarose gel and A260 absorbance measurement in a Nanodrop spectrophotometer (Thermo-Fisher Scientific Inc.). cDNA was prepared by reverse transcription of 1 µg of total RNA using high-fidelity reverse transcriptase and random hexamer supplied in a Prime ScriptTM Hi-fidelity RT-PCR kit. Quantitative real time PCR (qRT-PCR) analysis of the set of genes, including actin2, was performed using SYBR premix Ex-Taq in the ABI7300 (Applied Biosystems). Next, 4 µl of the 1/5-diluted cDNA was added to the 20 µl reaction mix containing 1X SYBR mix and 0.2 µM of each primer pairs (primer pairs for qRT-PCR are listed in Supplementary Table 1). Reactions were carried out in triplicates from two independent samples. The two-step PCR program consisted of 15 s of initial denaturation at 94°C followed by 40 cycles of 5 s at 94°C and 31 s at 60°C, with a final amplification for 1 min at 60°C. The amplification signals were collected at 60°C in each set of 40 cycles. Transcription levels were expressed as a fold change relative to the control samples, as calculated by the comparative (2−ΔΔCt) method [24].
ChIP-qPCR analysis
Isolation of chromatin and subsequent ChIP were performed using the Epiquick Plant ChIP Kit (Epigentek). Briefly, nuclei were isolated using sucrose gradient centrifugation and dissolved in nuclei lysis buffer. The chromatin was fragmented by sonication (Sonics and Materials Inc.) to an average fragment length of 500 bp (range of 200–700 bp) (see Supplementary Fig. 1). After centrifugation, the clear sonicated chromatin was collected from the supernatant and stored at −80°C for ChIP. Specific chromatin modifications were detected by immunoprecipitation using antibodies against H3K4me3, H3K9me2, H3K9ac, H3K27me3 and H3-core, according to the manufacturer's instructions (Epigentek). After column purification, immunoprecipitated DNA was diluted four times, and 4 µl was used for ChIP-qPCR reaction. For each gene, at least two types of primer pairs were designed: one pair from upstream promoter/untranslated (UTR) regions and another pair from gene body regions (see Supplementary Table 2). Quantitative PCR was performed as described for quantitative RT-PCR analysis. Each sample was assayed in duplicates. Two independent plant samples were used for ChIP-qPCR analysis of each gene or locus. Actin8_5’UTR and At4g03800_promoter regions were used as a positive control for the active (anti-H3K4me3) and the repressive chromatin marker (anti-H3K9me2), respectively. After normalization of qPCR data for the amount of input DNA, enrichment of chromatin due to immunoprecipitation was expressed as a percentage value of input [25]: input normalized IP (∆Ct) = (Ct[IP] − (Ct[input] − (log 2 input dilution factor))), and percentage input sites immunoprecipitated = (2−∆Ct) × 100%. In contrast, enrichment in site occupancy of each chromatin marker at specific genes or loci was calculated by the ΔΔCt method [24]: 2(−ΔΔCt[gamma ray/control]), where ΔΔCt[gamma ray/control] = (∆Ct[gamma ray]) − (∆Ct[control]). The ultimate fold change was expressed relative to actin8_5’UTR ChIP data. The results were statistically analyzed using Student's paired t test. A P-value of ≤0.05 was considered to be statistically significant.
DNA methylation analysis
DNA methylation levels of At1g31280 (AtAgo2), At2g30360 (AtCIPK11), At4g19130 (AtRPA1E), At5g20850 (AtRad51) and At5g24280 (AtGMI1) loci were determined by MethylC-Seq (bisulfite conversion followed by sequencing) analysis using genomic DNA from 4-week-old soil-grown plants. The growth condition and gamma irradiation for these plants were the same as described above. The methylC-seq analysis was performed using Illumina's HiSeq 2000 by BGI-Shenzhen, with some modifications in the methylC-seq library generation and high-throughput sequencing from the experimental procedures [26].
RESULTS
DNA damage repair genes showed upregulation of transcription after gamma irradiation
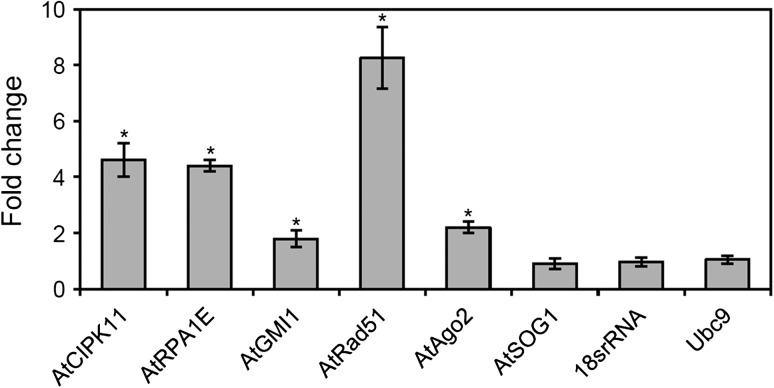
The transcript levels of six DNA damage repair genes, including three gamma-ray-responsive marker genes as identified previously [20], were investigated. About 2–8-fold induction of AtCIPK11, AtRPA1E, AtGMI1, AtRad51 and AtAgo2 transcription was detected by qRT-PCR analysis using actin2 as an endogenous control (Fig. 1). In contrast, the transcript level of AtSOG1, which is also known to be involved in DNA damage repair [27], essentially remained constant at 24 h after gamma irradiation (Fig. 1). Although AtSOG1 transcription was not induced by gamma rays, its protein activates the transcription of DSB repair genes via ATM-mediated phosphorylation [27, 28]. Thus, in the present study AtSOG1 was used as a negative control for gamma-ray-responsive transcription or for gamma-ray-responsive chromosomal loci associated with histone modifications.
Fig. 1.
Expression of DNA damage repair genes after gamma irradiation. Total RNA was isolated from 15-day-old seedlings and subjected to qRT-PCR analysis. Actin2 was used as an endogenous control gene to normalize for differences in the amount of total RNA. Error bars represent means ± SD (n = 4), and statistical significance of the values was evaluated by Student's paired t-test (*P < 0.05).
Site occupancy of H3K4me3 at the DNA damage repair genes after gamma irradiation generally increased
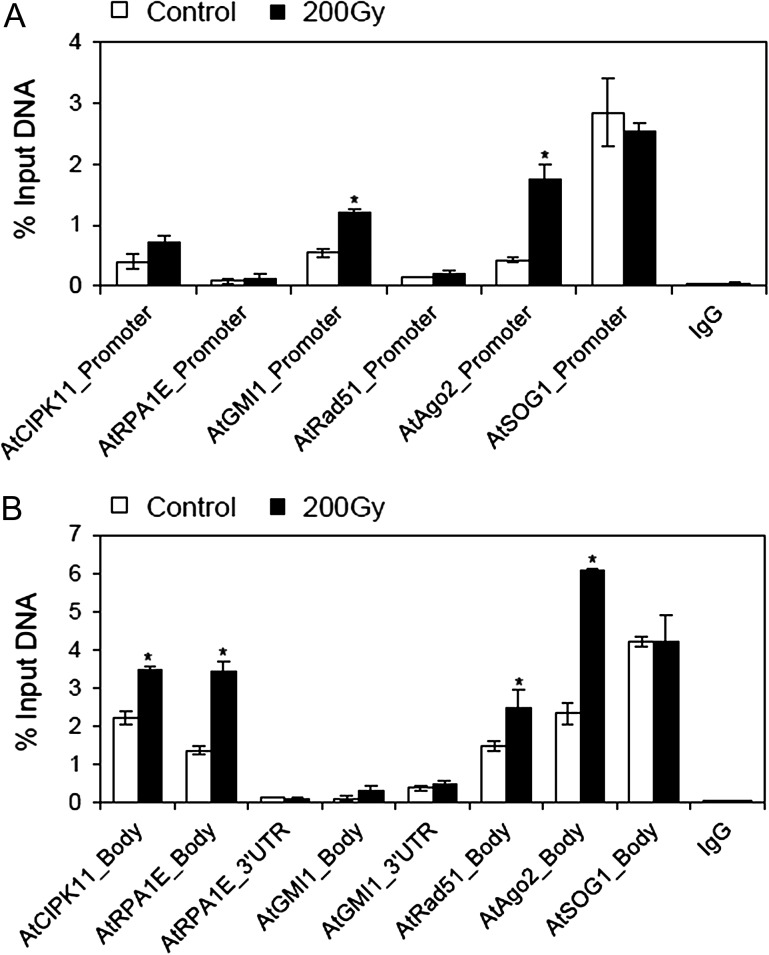
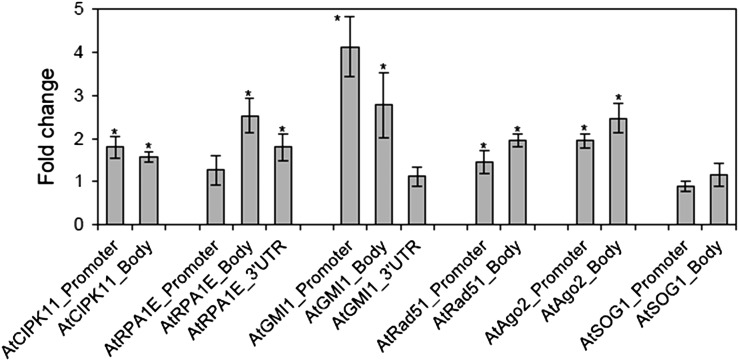
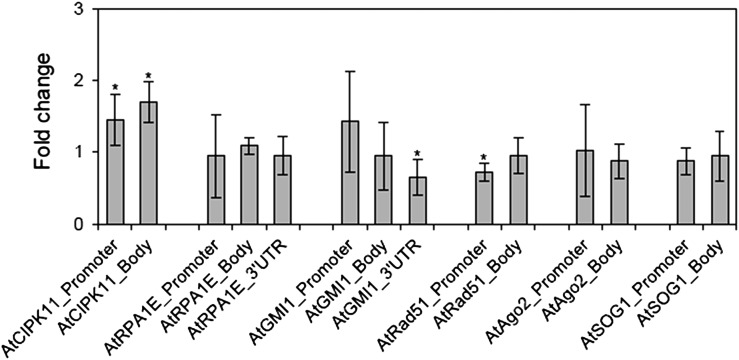
The active chromatin marker, H3K4me3, is known to be associated with transcriptionally active loci in chromosomes [29, 30]. Gamma rays influenced the association of H3K4me3 with different chromosomal loci for six DNA damage repair genes studied, showing a variation in the level of enrichment among the loci and among promoter, gene body, and UTR of each locus (Fig. 2). The percentage input values for the immunoprecipitation, using anti-H3K4me3 antibody, significantly increased in both the promoter and gene body regions of the AtAgo2 locus, whereas the AtGMI1 locus or the AtCIPK11, AtRPA1E and AtRad51 loci revealed significant enrichment of H3K4me3 in the promoter region or the gene body region, respectively (Fig. 2B). Fold change values in site occupancy of H3K4me3 after gamma irradiation were obtained by a multiple normalization using an IgG control antibody and non-immune serum (NIS) background for ChIP and using the actin8_5’UTR endogenous control gene for qPCR. The promoter, gene body, and/or 3’UTR regions of all the DNA damage repair genes (loci) tested, except AtSOG1, displayed a differential increase in the site occupancy of H3K4me3 after gamma irradiation (Fig. 3). For example, the site occupancy of H3K4me3 significantly increased in both the gene body regions of AtRPA1E and AtGMI1, while it increased ~4-fold only in the promoter region of AtGMI1, but not in that of AtRPA1E. For AtRad51 and AtAgo2, both the promoter and gene body regions showed a significant increase in the site occupancy of H3K4me3. Despite the highest percentage input values of H3K4me3 immunoprecipitation at the AtSOG1 locus, the site occupancy of H3K4me3 at this locus was not affected by gamma irradiation (Figs 2 and 3).
Fig. 2.
Changes in percentage input immunoprecipitation of the H3K4me3 marker at DNA damage repair genes after gamma irradiation. (A) and (B) represent percentage input immunoprecipitation of the H3K4me3 marker at promoter and gene body or untranslated (UTR) regions of DNA damage repair genes, respectively. ‘Gene body’ is a transcription unit including both exons and introns. Error bars represent means ± SD (n = 4), and the statistical significance of the values was evaluated by Student's paired t-test (*P < 0.05).
Fig. 3.
Changes in the site occupancy of H3K4me3 at DNA damage repair genes after gamma irradiation. Fold change was calculated by the ΔΔCt method described in the Materials and Methods section using actin8_5’UTR as a control. ‘Gene body’ is a transcription unit including both exons and introns. Error bars represent means ± SD (n = 4), and the statistical significance of the values was evaluated by Student's paired t-test (*P < 0.05).
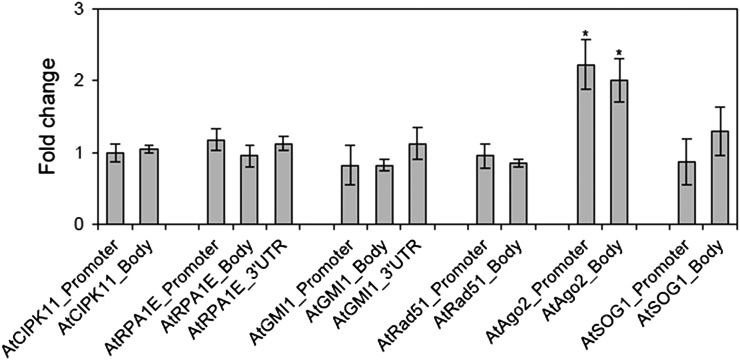
Another active chromatin marker, H3K9ac, showed enrichment only at the AtAgo2 locus after gamma irradiation
Several studies have reported colocalization of H3K4me3 and H3K9ac markers and suggested a cooperative interaction between them [13, 31]. Interestingly, after gamma irradiation we observed significantly higher percentage input immunoprecipitation and fold change in the site occupancy of H3K9ac only at both the promoter and gene body regions of the AtAgo2 locus (Fig. 4 and see Supplementary Fig. 2). No other loci, including AtSOG1, showed any significant enrichment of H3K9ac. However, the comparatively higher percentage input values for the immunoprecipitation using the anti-H3K9ac antibody indicated that DNA damage repair genes may be associated more with H3K9ac compared with other histone markers, irrespective of gamma irradiation (see Supplementary Fig. 2).
Fig. 4.
Changes in the site occupancy of H3K9ac at DNA damage repair genes after gamma irradiation. Fold change was calculated by the ΔΔCt method described in the Materials and Methods section using actin8_5’UTR as a control. ‘Gene body’ is a transcription unit including both exons and introns. Error bars represent means ± SD (n = 4), and the statistical significance of the values was evaluated by Student's paired t-test (*P < 0.05).
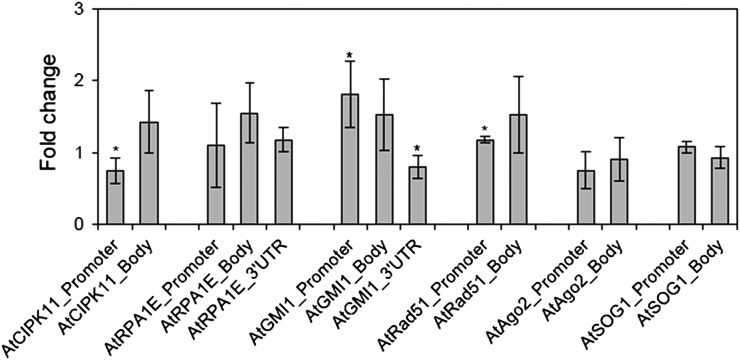
Site occupancy of H3K9me2 was not universally changed at the DNA damage repair genes after gamma irradiation
To confirm the enhanced interactions of the DNA damage repair genes with active chromatin markers (H3K4me3 and H3K9ac) after gamma irradiation, the fold change in site occupancy of an inactive chromatin marker (H3K9me2) was also evaluated in the promoter, gene body, and/or 3’UTR regions of those genes. The 3’UTR region of AtGMI1 and the promoter region of AtRad51 displayed significant reduction in the site occupancy of H3K9me2, thus correlating with the increased transcription levels of the genes, as shown in Fig. 1 (Fig. 5). In contrast, the AtCIPK11 locus showed a slight but significant increase in the site occupancy of H3K9me2 at both the promoter and gene body regions. These changes were not significant at the AtRPA1E and AtAgo2 loci. Additionally, the site occupancy of H3-core (histone occupancy) was compared among the tested loci and various regions of each locus. The site occupancy of H3-core significantly decreased in the promoter region of AtCIPK11 and the 3’UTR region of AtGMI1, while it increased in the promoter regions of AtGMI1 and AtRad51 (Fig. 6). Therefore, it is suggested that active histone markers such H3K4me3 and H3K9ac are more important for transcription of the DNA damage repair genes in plants than other histone markers such as H3K9me2 and H3-core.
Fig. 5.
Changes in the site occupancy of H3K9me2 at DNA damage repair genes after gamma irradiation. Fold change was calculated by the ΔΔCt method described in the Materials and Methods section using actin8_5’UTR as a control. ‘Gene body’ is a transcription unit including both exons and introns. Error bars represent means ± SD (n = 4), and the statistical significance of the values was evaluated by Student's paired t-test (*P < 0.05).
Fig. 6.
Changes in the site occupancy of H3-core at DNA damage repair genes after gamma irradiation. Fold change was calculated by the ΔΔCt method described in the Materials and Methods section using actin8_5’UTR as a control. ‘Gene body’ is a transcription unit including both exons and introns. Error bars represent means ± SD (n = 4), and the statistical significance of the values was evaluated by Student's paired t-test (*P < 0.05).
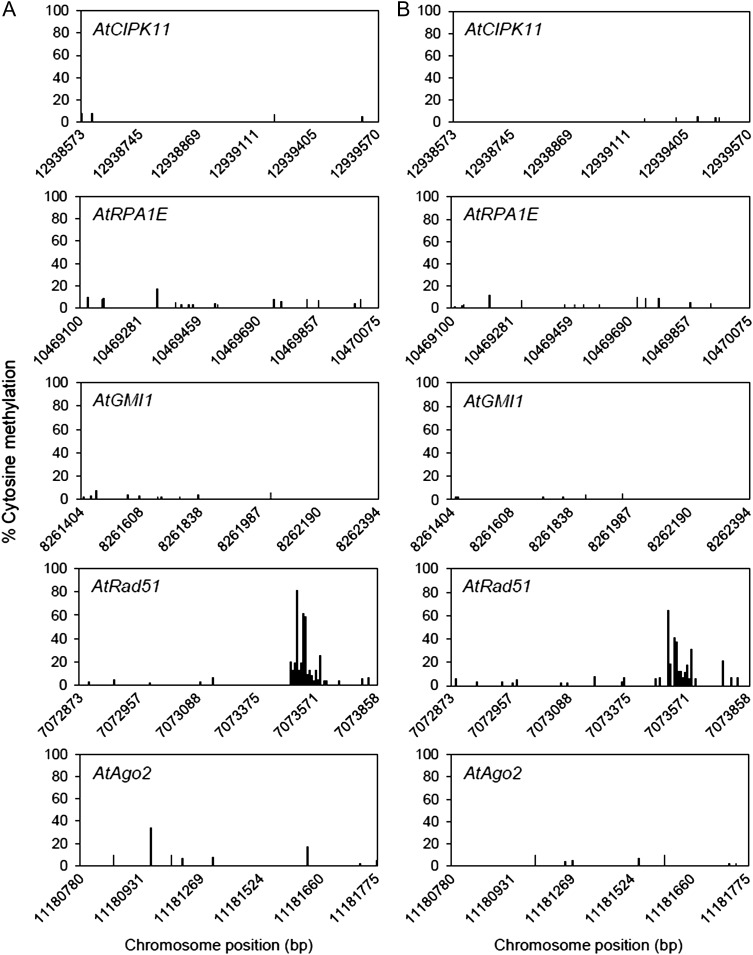
Cytosine methylation levels at the promoter regions of the DNA damage repair genes did not significantly change after gamma irradiation
DNA methylation affects transcription [32], and histone methylation or acetylation is associated with DNA methylation [33, 34]. Therefore, we investigated cytosine methylation levels in the promoter regions of the five gamma-ray-inducible DNA damage repair genes in the genome-wide MethylC-Seq data, which were obtained via bisulfite conversion followed by sequencing. The cytosine methylation levels in the promoter regions of the AtCIPK11, AtRPA1E, AtGMI1, AtRad51 and AtAgo2 loci, which are inducible in response to gamma rays, remained almost unaffected after gamma irradiation (Fig. 7). Moreover, these promoter regions did not display substantial changes in cytosine methylation. These results suggest that change in DNA methylation cannot be a key player in the gamma-ray-inducible chromatin remodelling associated with transcriptional activation of the DNA damage repair genes.
Fig. 7.
Changes in cytosine methylation of putative promoter regions of DNA damage repair genes after gamma irradiation. Approximately 1-kb sequences upstream from the transcription start site of each gene were extracted from the genome-wide methylC-seq data and used as a putative promoter region for cytosine methylation analysis. This promoter region includes the sequences subjected to ChIP-qPCR analysis in Figs 1–6. (A) and (B) represent the control and gamma-irradiated groups, respectively.
DISCUSSION
It has been previously reported that several genes, such as AtRad51, AtAgo2, AtCIPK11, AtRPA1E and AtGMI1, are gamma-ray-inducible and play an important role in the repair of DNA damage caused by gamma irradiation (see Supplementary Table 3) [18, 20, 35]. Here we demonstrate that transcriptions of these genes were substantially induced by gamma irradiation and that the induction could in part be associated with histone modifications. Histone modifications include phosphorylation, acetylation, methylation, ubiquitination, ADP-ribosylation and sumoylation [36, 37]. Although there is little information available on histone modifications in plants upon exposure to ionizing radiation, a H2A variant H2AX, is known to be phosphorylated and to work as a major sensor for DSB sites in chromatin [6]. The present work demonstrates significant enrichment of the active chromatin marker H3K4me3 at the five DNA damage repair genes or loci after gamma irradiation (Figs 2 and 3). These results indicate that the enrichment of H3K4me3 is important for gamma-ray-inducible transcription at these specific loci and thus for DNA damage (especially DSB) repair. It has also been reported that phosphorylated H2AX (γH2AX) foci preferentially occur in the actively transcribing euchromatic regions (enriched with H3K4me3) following gamma irradiation [38]. Since the H3K4me3 marker is neutral in charge, it cannot directly perturb chromatin structure [36]. Instead, this marker increased accessibility of the chromatin template to the transcriptional machinery by recruiting CHD1- and BPTF-like ATP-dependent chromatin remodelling factors [39]. In addition, the H3K4me3 marker may help to maintain the chromatin in the acetylated state by interrupting the binding of the nucleosome remodelling and deacetylase (NuRD) complex to the H3 N-terminal tail [40]. Based on other relevant data in yeast, it has also been suggested that H3K4me3 could be involved in the recruitment of chromatin remodellers and maintenance of genome stability after DNA damage [9]. Nevertheless, the AtSOG1 gene showed neither enhanced transcription nor enrichment of the H3K4me3 marker after gamma irradiation. Thus, this gene was used as a negative control to interpret the relationship between the locus-specific histone modification and enhancement of transcription after gamma irradiation. The AtSOG1 acts as a transcription factor and its activity depends on the ATM-mediated phosphorylation of the C-terminal end [28].
Another active chromatin marker, H3K9ac, is mainly associated with the gene body regions of actively transcribing loci [41]. It is therefore to be noted that all the DNA damage repair genes or loci tested in this study except AtGMI1 generally showed more occupancy of the H3K9ac at the gene body regions than at the promoter regions in both control and irradiated samples (see Supplementary Fig. 2). Moreover, occupancy of the H3K9ac markers in control samples implies that the genes and loci are transcriptionally active, even under normal conditions prior to gamma irradiation. However, since the only significant fold change in site occupancy of the H3K9ac markers after gamma irradiation was found at the AtAgo2 locus, the H3K9ac enrichment may play an important role in gamma-ray-inducible transcription of AtAgo2. Both enrichment of the H3K4me3 and H3K9ac markers at the AtAgo2 locus can be associated with plant homeo domain (PHD)–containing proteins, which may facilitate chromatin remodelling by recruiting histone acetyl transferase to chromatin [42, 43]. The PHD-containing proteins recognize the H3K4me3 marker and act as an interpreter for this code [44].
Nucleosome density or nucleosome occupancy is decreased and the chromatin structure relaxed during transcriptional activation of a genomic region [45, 46]. Nucleosome occupancy can be measured through ChIP assay using H3 C-terminal antibody. We have demonstrated that the site occupancy of the H3 core was significantly reduced at the promoter region of AtCIPK11 and the 3’UTR region of AtGMI1 after gamma irradiation. Similarly, histone occupancy decreases at the promoter regions of the COR15A and ATGOLS3 genes in response to cold stress [14]. The inactive chromatin marker, H3K9me2, is negatively correlated with transcriptionally active loci [41]; however, the correlation was observed only at the 3’UTR region of AtGMI1 and the promoter region of AtRad51. The site occupancy of the H3 core or the H3K9me2 marker appears to be less affected by gamma rays than that of the H3K4me3, depending on the genes or loci tested.
Plants have an integral association between chromomethylase 3 (CMT 3) proteins and H3K9me2 markers in chromatin, which recruit each other through the formation of a self-reinforcing loop for the maintenance of heterochromatic regions, including the promoter regions of repressed genes or transposable elements [47, 48]. However, the significant changes in the H3K9me2 occupancy at the promoter regions of the AtCIPK11 and AtRad51 loci after gamma irradiation did not correlate with their cytosine methylation levels. Although these genes were substantially and reliably induced 24 h after gamma irradiation of 200 Gy at a dose rate of 50 Gy h−1 in both the MS-medium-grown 15-day-old seedlings (Fig. 1) and the soil-grown 28-day-old seedlings [20], their methylation was maintained at a constant very low level before and after gamma irradiation (Fig. 7). It is thus unlikely that the dynamic changes in the site occupancy of active and inactive chromatin markers in response to gamma irradiation are closely correlated with DNA methylation. Lack of relationship between the genome-wide transcriptional changes and DNA methylation levels after gamma irradiation was also demonstrated in an earlier study [20]. Based collectively on the above information, we suggest that histone modifications could contribute to the transcriptional activation of the five DNA damage repair genes, possibly by inducing changes in the chromatin structure without causally affecting the DNA methylation. It is expected that the hypothesis advanced here may also be applicable to other gamma-ray-responsive genes.
In conclusion, our work demonstrated that gamma irradiation induced dynamic histone modifications in Arabidopsis seedlings. Among these, methylation at lys 4 of H3 (H3K4me3) was generally found at the gamma-ray-inducible loci involved in DNA repair, while acetylation at lys 9 of H3 (H3K9ac) was enriched only at the AtAgo2 locus. These results imply that induction of gamma-ray-mediated histone modifications is locus-specific. Future studies should target unravelling kinetics (time scale) and mechanisms underlying such specific chromatin modifications in response to gamma rays.
SUPPLEMENTARY DATA
Supplementary data are available at Journal of Radiation Research online.
FUNDING
This research was supported by the Nuclear R & D Program funded by the Ministry of Science, ICT and Future Planning (MSIP), Republic of Korea.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
REFERENCES
- 1.Boyko A, Zemp F, Filkowski J, et al. . Double strand break repair in plants is developmentally regulated. Plant Physiol 2006;141:488–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rogakou EP, Boon C, Redon C, et al. . Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol 1999;146:905–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vidanes GM, Bonilla CY, Toczyski DP.. Complicated tails: histone modifications and the DNA damage response. Cell 2005;121:973–6. [DOI] [PubMed] [Google Scholar]
- 4.Widlak P, Pietrowska M, Lanuszewska J.. The role of chromatin proteins in DNA damage recognition and repair. Histochem Cell Biol 2006;125:119–26. [DOI] [PubMed] [Google Scholar]
- 5.Williamson EA, Wray JW, Bansal P, et al. . Overview for the histone codes for DNA repair. Prog Mol Biol Transl Sci 2012;110:207–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunt CR, Ramnarain D, Horikoshi N, et al. . Histone modifications and DNA double-strand break repair after exposure to ionizing radiations. Radiat Res 2013;179:383–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma GG, So S, Gupta A, et al. . MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol Cell Biol 2010;30:3582–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanders SL, Portoso M, Mata J, et al. . Methylation of histone H4 lysine 20 controls recruitment of Crb2 to site of DNA damage. Cell 2004;119:603–14. [DOI] [PubMed] [Google Scholar]
- 9.Faucher D, Wellinger RJ.. Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genet 2010;6:e1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikura T, Tashiro S, Kakino A, et al. . DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol Cell Biol 2007;27:7028–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez-Capetillo O, Mahadevaiah SK, Celeste A, et al. . H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev Cell 2003;4:497–508. [DOI] [PubMed] [Google Scholar]
- 12.Millar CB, Grunstein M.. Genome-wide patterns of histone modifications in yeast. Nat Rev Mol Cell Biol 2006;7:657–66. [DOI] [PubMed] [Google Scholar]
- 13.Kim JM, To TK, Ishida J, et al. . Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol 2008;49:1580–8. [DOI] [PubMed] [Google Scholar]
- 14.Kwon CS, Lee D, Choi G, et al. . Histone occupancy–dependent and independent removal of H3K27 trimethylation at cold-responsive genes in Arabidopsis. Plant J 2009;60:112–21. [DOI] [PubMed] [Google Scholar]
- 15.Sokol A, Kwiatkowska A, Jerzmanowski A, et al. . Upregulation of stress-inducible genes in tobacco and Arabidopsis cells in response to abiotic stresses and ABA treatment correlates with dynamic changes in histone H3 and H4 modifications. Planta 2007;227:245–54. [DOI] [PubMed] [Google Scholar]
- 16.Tsuji H, Saika H, Tsutsumi N, et al. . Dynamic and reversible changes in histone H3-Lys4 methylation and H3 acetylation occurring at submergence-inducible genes in rice. Plant Cell Physiol 2006;47:995–1003. [DOI] [PubMed] [Google Scholar]
- 17.Nagata T, Yamada H, Du Z, et al. . Microarray analysis of genes that respond to γ-irradiation in Arabidopsis. J Agric Food Chem 2005;53:1022–30. [DOI] [PubMed] [Google Scholar]
- 18.Culligan KM, Robertson CE, Foreman J, et al. . ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J 2006;48:947–61. [DOI] [PubMed] [Google Scholar]
- 19.Kim DS, Kim JB, Goh EJ, et al. . Antioxidant response of Arabidopsis plants to gamma irradiation: genome-wide expression profile of the ROS scavenging and signal transduction pathways. J Plant Physiol 2011;168:1960–71. [DOI] [PubMed] [Google Scholar]
- 20.Kim JH, Kim JE, Lee MH, et al. . Integrated analysis of diverse transcriptomic data from Arabidopsis reveals genetic markers that reliably and reproducibly respond to ionizing radiation. Gene 2013;518:273–9. [DOI] [PubMed] [Google Scholar]
- 21.Aklilu BB, Soderquist RS, Culligan KM.. Genetic analysis of the Replication Protein A large subunit family in Arabidopsis reveals unique and overlapping roles in DNA repair, meiosis and DNA replication. Nucleic Acids Res 2014;42:3104–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Böhmdorfer G, Schleiffer A, Brunmeir R, et al. . GMI 1, a structural-maintenance-of-chromosomes-hinge domain-containing protein, is involved in somatic homologous recombination in Arabidopsis. Plant J 2011;67:420–33. [DOI] [PubMed] [Google Scholar]
- 23.Wei W, Ba Z, Gao M, et al. . A role for small RNAs in DNA double-strand break repair. Cell 2012;149:101–12. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD.. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- 25.Haring M, Offermann S, Danker T, et al. . Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 2007;3:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lister R, O'Malley RC, Tonti-Filippini J, et al. . Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 2008;133:523–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshiyama K, Conklin PA, Huefner ND, et al. . Supressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc Natl Acad Sci U S A 2009;106:12843–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshiyama KO, Kobayashi J, Ogita N, et al. . ATM-mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis. EMBO Rep 2013;14:817–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernstein BE, Humphrey EL, Erlich RL, et al. . Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci U S A 2002;99:8695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santos-Rosa H, Schneider R, Bannister AJ, et al. . Active genes are tri-methylated at K4 of histone H3. Nature 2002;419:407–11. [DOI] [PubMed] [Google Scholar]
- 31.Malapeira J, Khaitova LC, Mas P.. Ordered changes in histone modifications at the core of the Arabidopsis circadian clock. Proc Natl Acad Sci U S A 2012;109:21540–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le TN, Miyazaki Y, Takuno S, et al. . Epigenetic regulation of intragenic transposable elements impacts gene transcription in Arabidopsis thaliana. Nucleic Acids Res 2015;43:3911–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cedar H, Bergman Y.. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 2009;10:295–304. [DOI] [PubMed] [Google Scholar]
- 34.Qian W, Miki D, Zhang H, et al. . A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science 2012;336:1445–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao M, Wei W, Li MM, et al. . Ago2 facilitates Rad51 recruitment and DNA double-strand break repair by homologous recombination. Cell Res 2014;24:532–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bannister AJ, Kouzarides T.. Regulation of chromatin by histone modifications. Cell Res 2011;21:381–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Millar CB, Grunstein M.. Genome-wide patterns of histone modifications in yeast. Nat Rev Mol Cell Biol 2006;7:657–66. [DOI] [PubMed] [Google Scholar]
- 38.Vasireddy RS, Karagiannis TC, El-Osta A.. γ-radiation-induced γH2AX formation occurs preferentially in actively transcribing euchromatic loci. Cell Mol Life Sci 2010;67:291–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruthenberg AJ, Allis CD, Wysocka J.. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 2007;25:15–30. [DOI] [PubMed] [Google Scholar]
- 40.Zegerman P, Canas B, Pappin D, et al. . Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) repressor complex. J Biol Chem 2002;277:11621–4. [DOI] [PubMed] [Google Scholar]
- 41.Zhou J, Wang X, He K, et al. . Genome-wide profiling of histone H3 lysine 9 acetylation and dimethylation in Arabidopsis reveals correlation between multiple histone marks and gene expression. Plant Mol Biol 2010;72:585–95. [DOI] [PubMed] [Google Scholar]
- 42.Pena PV, Davrazou F, Shi X, et al. . Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 2006;442:100–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi X, Hong T, Walter KL, et al. . ING2 PHD domain links histone H3 lysine 4 methylation to active gene expression. Nature 2006;442:96–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee WY, Lee D, Chung WI, et al. . Arabidopsis ING and Alfin1-like protein families localize to the nucleus and bind to H3K4me3/2 via plant homeodomain fingers. Plant J 2009;58:511–24. [DOI] [PubMed] [Google Scholar]
- 45.Clark DJ, Felsenfeld G.. Formation of nucleosomes on positively supercoiled DNA. EMBO J 1991;10:387–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eberharter A, Becker PB.. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep 2002;3:224–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson LM, Bostick M, Zhang X, et al. . The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol 2007;17:379–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Law JA, Jacobsen SE.. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 2010;11:204–20. [DOI] [PMC free article] [PubMed] [Google Scholar]