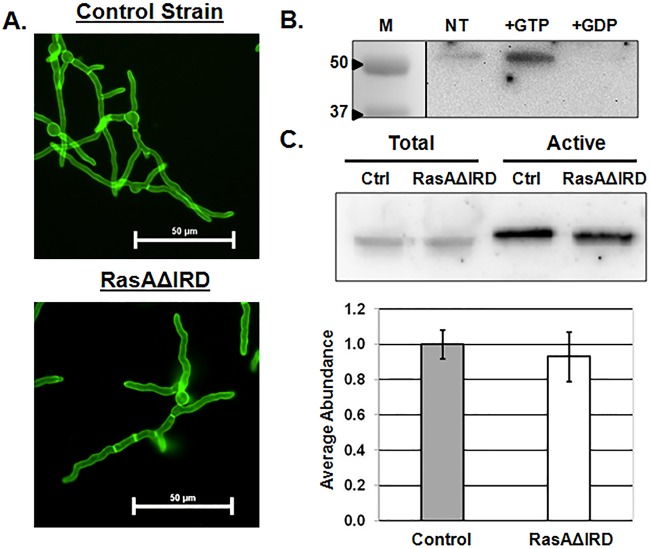
FIG 5 .
RasA localization and activation are not affected by IRD truncation. (A) Representative fluorescence micrographs of the control strain and RasAΔIRD. AMM broth cultures of each strain were incubated at 37°C until approximately equal hyphal lengths were achieved for each strain. Cultures were then mounted for fluorescence microscopy, and images were acquired using identical exposure settings for both strains. (B) Ras activation assay performed on lysate from the control strain comparing the amount of total Ras bound under no treatment (NT), after GTP preloading (+GTP), and after GDP preloading (+GDP). GFP-fused RasA is detected at 51 kDa using an anti-Ras antibody. The control assay was repeated twice to ensure the ability of RasA to bind the Raf1 RBD-agarose bead and to appropriately respond to activation (+GTP) and inactivation (+GDP). Lane M, molecular mass (kilodaltons) marker lane.(C, top panel) Western blot analysis of total RasA detected in lysates from each strain (Total) and the amount of active RasA detected by Raf1-RBD pulldown (Active). The gel image is representative of three independent experiments. Ctrl, control. (C, bottom panel) Quantification of active/total Ras ratios. Western blot bands were quantified by densitometry analysis, and the average signal abundance ratio of detectable active RasA to total RasA was calculated. Data are presented as an average from three replicates ± standard deviation. No statistical difference was noted in Ras activation levels between the control and RasAΔIRD mutant.