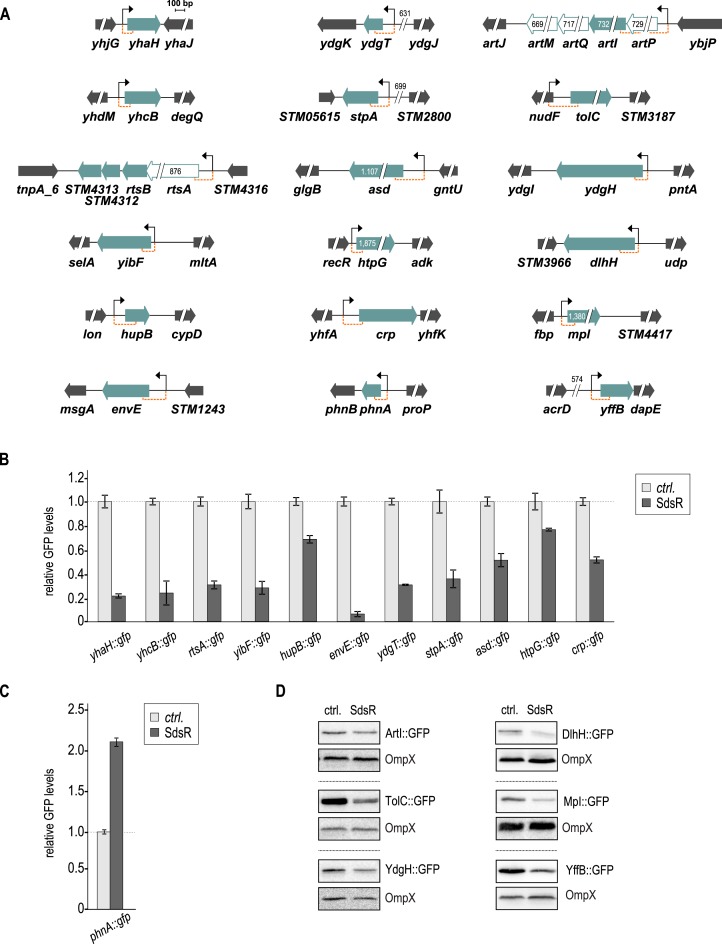
Figure 1.
Genomic context of SdsR target genes and validation with gfp reporter fusions. (A) Genomic location and flanking regions of confirmed SdsR targets. Target genes are shown as filled green arrows; genes within the same transcriptional unit as validated targets are represented as open green arrows. All genes are drawn to scale; where appropriate, numbers indicate fragment sizes. Transcriptional start sites (as determined by (54)) are marked by a black arrow, and sequence stretches represented in gfp reporter fusions are indicated by orange brackets. (B) Repression of target genes by SdsR. Salmonella ΔsdsR cells carrying the indicated gfp reporter fusion in combination with either a control, or a plasmid expressing Salmonella SdsR were grown to early stationary phase (OD600 of 2), fixed with PFA, and GFP fluorescence was determined by flow cytometry. For each GFP-fusion, fluorescence levels in the presence of the control plasmid were set to 1, and relative changes were determined for cells expressing SdsR. GFP levels were calculated from three biological replicates; error bars indicate the standard deviation. Detailed descriptions of all plasmids are provided in Supplementary Table S2. (C) Positive regulation of phnA::gfp in the presence of SdsR. Experimental procedure as in (B). (D) Validation of non-fluorescent target fusions by western blot analysis. Protein samples were collected from Salmonella ΔsdsR cells grown to early stationary phase (OD600 of 2) and carrying the indicated gfp reporter fusion in combination with either a control or a plasmid expressing Salmonella SdsR. OmpX was probed as loading control. A quantification of these results is provided in Supplementary Table S4.