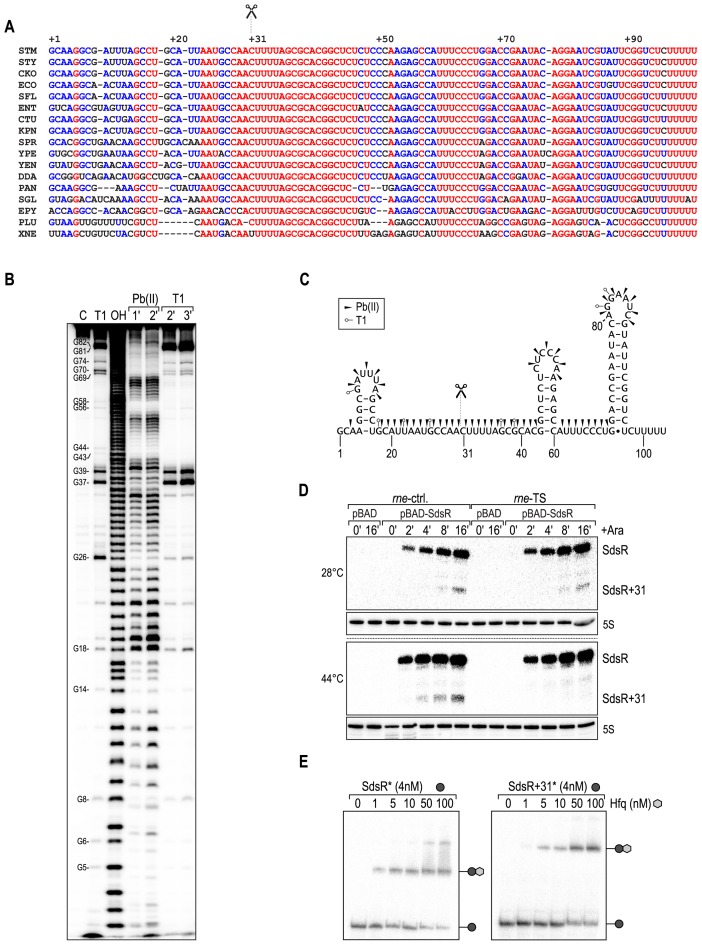
Figure 2.
SdsR sRNA is processed by RNase E, and associates with Hfq. (A) Multiple alignment of SdsR sRNA sequences of diverse enterobacteria. Fully, partially and poorly conserved nucleotides are indicated in red, blue and black, respectively. Scissors mark the RNase E processing site. Abbreviations correspond to the following species: STM, Salmonella Typhimurium; STY, Salmonella typhi; CKO, Citrobacter koseri; ECO, Escherichia coli; SFL, Shigella flexneri; ENT, Enterobacter sp. 638; CTU: Cronobacter turicensis; KPN, Klebsiella pneumoniae; SPR, Serratia proteamaculans; YPE, Yersinia pestis; YEN, Yersinia enterocolitica; DDA, Dickeya dadantii; PAN, Pantoea ananatis; SGL, Sodalis glossinidius; EPY, Erwinia pyrifoliae; PLU, Photorhabdus luminescens; XNE, Xenorhabdus nematophila. (B) Determination of the SdsR structure by in vitro probing. 5′ end-labelled SdsR was subjected to Pb(II) acetate (lanes 4 and 5) or RNase T1 cleavage (lanes 6 and 7), and reactions were stopped at indicated time-points. RNase T1 and alkaline (OH) ladders were used to map cleaved fragments. Positions of G-residues are indicated. (C) Secondary structure of SdsR. Cleavage sites as determined in (B) are indicated by arrowheads (Pb(II) acetate) or open circles (T1). Scissors mark the RNase E cleavage site. (D) SdsR processing is dependent on RNase E. Salmonella rne-TS and its isogenic control strain carrying either pBAD-SdsR or control plasmid pBAD were grown at the permissive temperature of 28°C to early stationary phase (OD600 of 1), when cultures were split, and growth was continued for 30 min at either 28°C, or 44°C to inactivate RNase E. Expression of SdsR was induced by the addition of l-arabinose, and RNA harvested at the indicated time-points was analyzed on Northern blots using a SdsR-specific probe. 5S rRNA served as loading control. (E) Full-length and processed SdsR associate with Hfq. Electrophoretic mobility shift assay (EMSA) of in vitro synthesized 5′-end-labelled SdsR or SdsR+31 RNAs (4 nM) in the presence of increasing concentrations of Hfq protein as indicated.