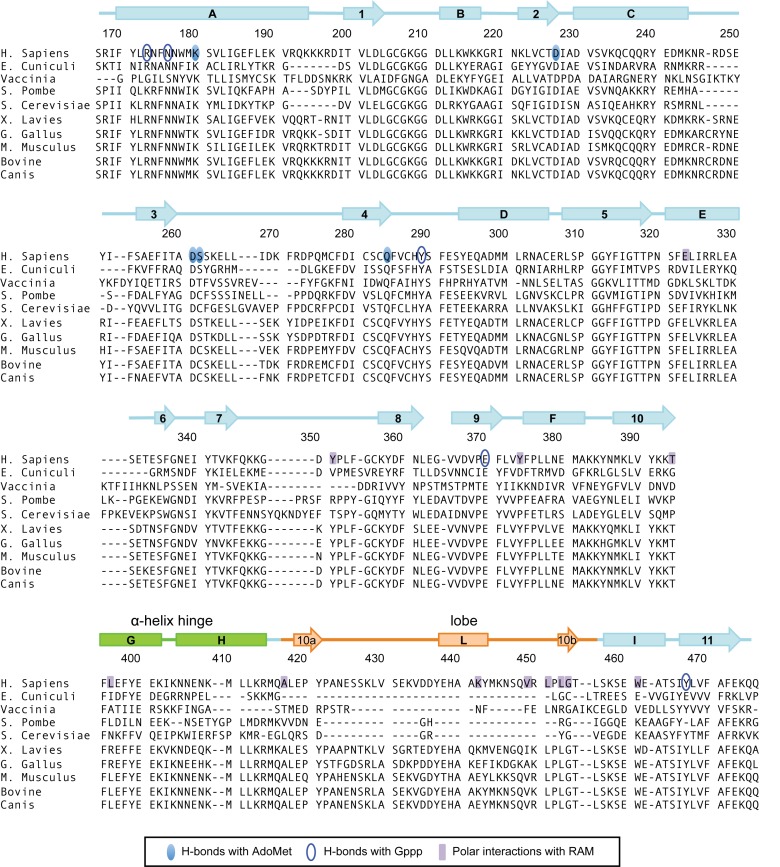
Figure 2.
Sequence alignments and secondary structure element attribution for human RNMT. Amino acid sequence alignment using EMBL-EBI Clustal Omega between RNMT orthologs from a selection of eukaryotic organisms. Secondary structure attribution for RNMT (PDB: 5E8J) was performed with DSSP. The nomenclature of structural features shown has been transcribed from Fabrega et al. (15). Previously unidentified features include strands 10a, 10b and helix L, which lies within the lobe structure (orange) found to be conserved amongst vertebrates. Helices H and G form the α-helix hinge (green). Amino acids involved in polar interaction with AdoMet, Gppp and RAM are highlighted.