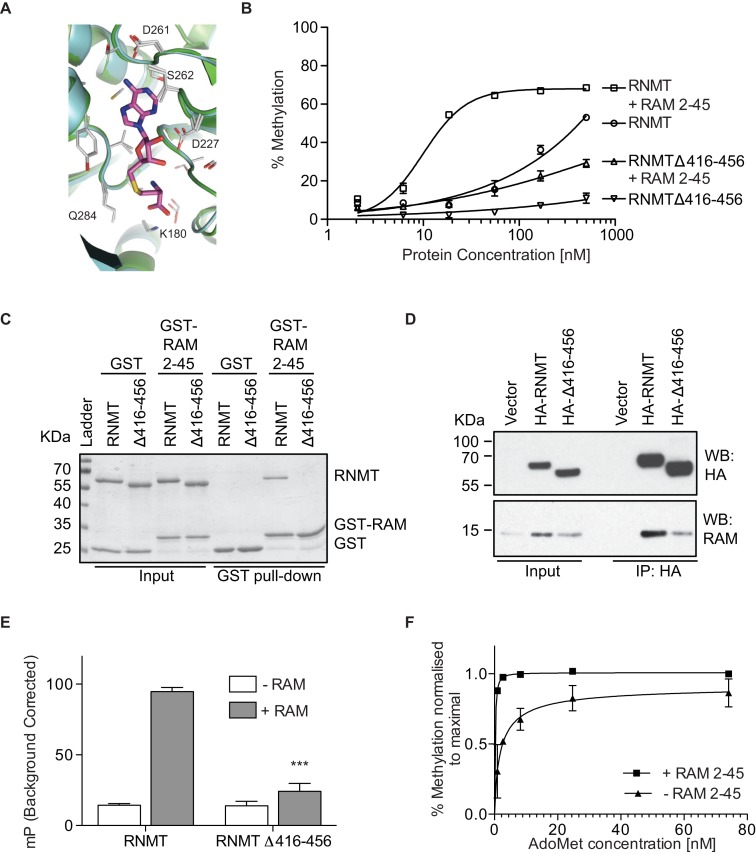
Figure 3.
RAM increases AdoMet recruitment by RNMT. (A) Alignment of residues involved in AdoHcy binding for RNMT 165–476–RAM 2–45 (cyan) and RNMT 165–476 Δ416–456 monomer (green). Residues from both structures involved in polar interactions are shown as sticks. (B) Methyltransferase activity assays performed with titrations of RNMT and RNMT Δ416-456 in presence or absence of RAM (n = 3). (C) GST-pulldown of RNMT and RNMT Δ416–456 with GST-RAM 2–45. GST provides the negative control. (D) Co-immunoprecipitation followed by Western blot analysis on lysates from cells transiently transfected with vector only, HA-RNMT and HA-RNMT Δ416–456. (E) Maximal polarisation for SAMFP probe binding to RNMT and RNMT Δ416-456 in absence or presence of RAM 2–45. 0.1 mM GpppG present throughout (n = 3). P-values are relative to RNMT controls where *** represents P < 0.001. (F) Methyltransferase activity assays performed with 100 nM RNMT in presence or absence of equimolar RAM 2–45 and an AdoMet titration (n = 2). Values in B, E and F represent mean ± S.D. Panels C and D represent two or more independent experiments.