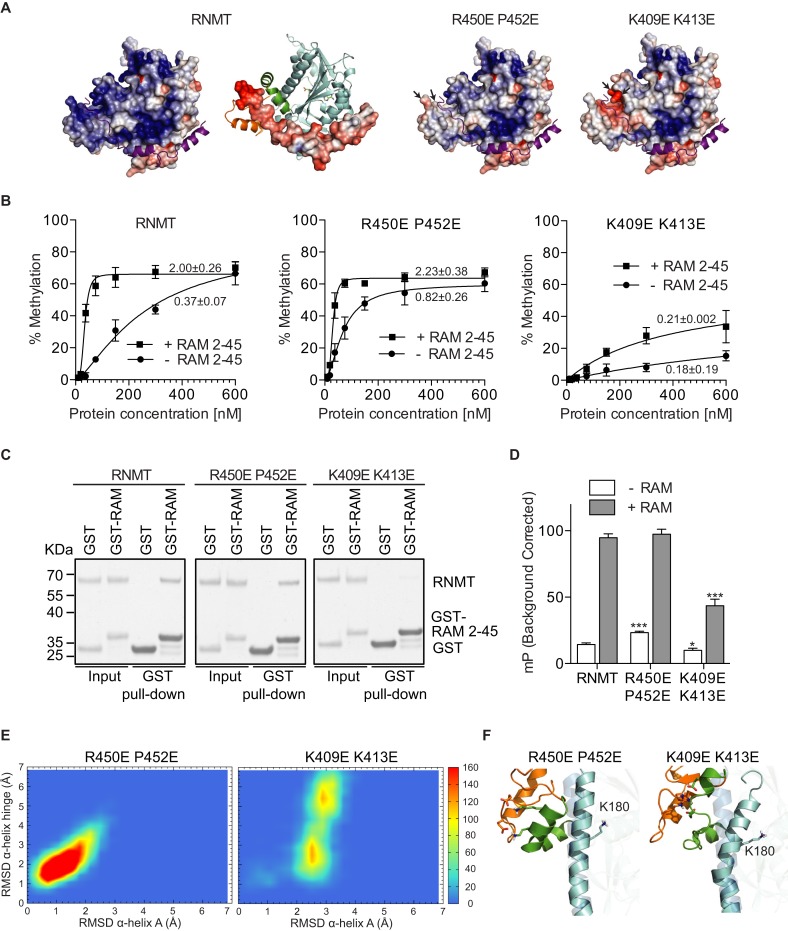
Figure 5.
Reversing RNMT lobe charge increases enzymatic activity. (A) Electrostatic potential as calculated by APBS from the crystal structure of RNMT-RAM. Positive charge is depicted in blue and negative charge in red (–5/+5 KbT/e). Predicted alterations in charge due to amino acid substitutions on the lobe surface (P452E R450E) or surface facing the lobe (K409E K413E) are demonstrated. (B) Methyltransferase activity assays performed on RNMT, RNMT R450E P452E and RNMT K409E K413E in absence or presence of RAM (n = 3). Activity was calculated as % methylation per ng RNMT in a 5-minute reaction. Maximal activity over the titration is reported. (C) GST-pulldown of RNMT, RNMT R450E P452E and RNMT K409E K413E with GST-RAM 2–45. GST provides the negative control. (D) Maximal polarization for SAMFP probe binding to RNMT, RNMT P450E R452E and RNMT K409E K413E in absence or presence of RAM 2–45. (E) Heatmaps displaying backbone RMSD of the hinge (residues 395–416) versus RMSD of helix A (residues 341–345) as calculated from 60ns high-temperature MD trajectories of RNMT R452E R450E and RNMT K409E K413E mutants. The color scale represents the number of snapshots in each ‘bin’. (F) Representative snapshots corresponding to the most sampled configuration in panel E. Helix A is shown in blue, hinge in green and lobe in orange. High-transparency image of helix A in the background represents the position in RNMT–RAM crystal structure. Values in panels B and D represent mean ± SD (n = 3). P-values in panel D are relative to RNMT controls where * represents P < 0.05 and *** represents P < 0.001.