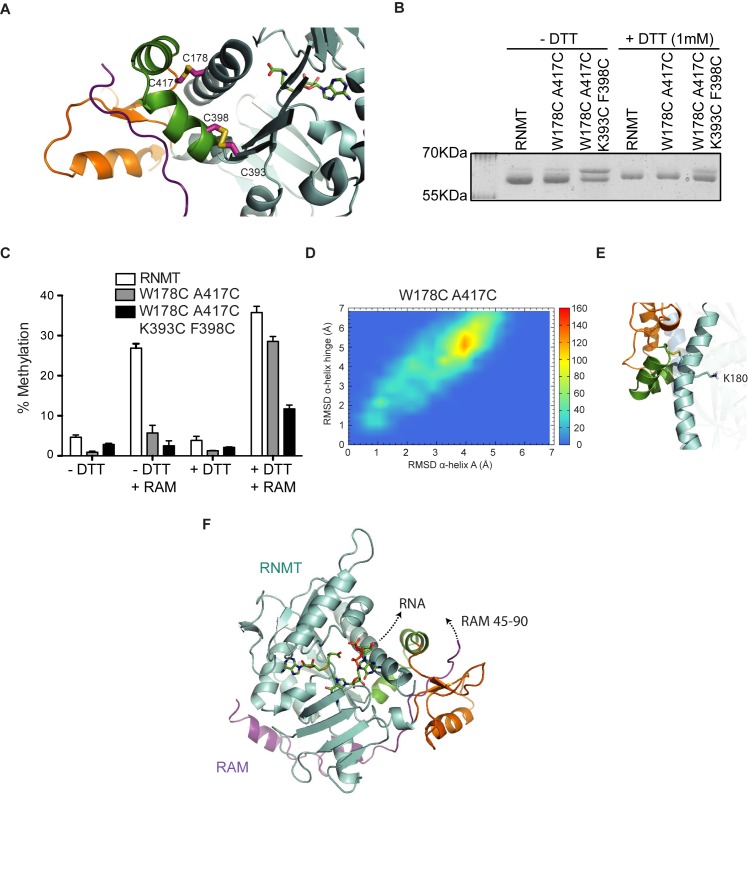
Figure 6.
Restraining the RNMT α-helix hinge ablates its catalytic activity. (A) Predicted structures for amino acid substitutions introducing disulphide bridges designed to constrain the alpha helix hinge (green) to helix A. Introduced disulphide bridges are shown as sticks. (B) Coomassie stained SDS PAGE gels resolving RNMT, RNMT W178C A417C (single-bridge mutant) and RNMT W178C A417C K393C F398C (double-bridge mutant) in the absence and presence of 1 mM DTT. (C) Methyltransferase activity assays performed on 100nM RNMT, RNMT W178C A417C and RNMT W178C A417C K393C F398C, with or without RAM 2–45 with or without 1mM DTT. (D) Heatmaps displaying the backbone RMSD of the hinge (residues 395–416) versus RMSD of helix A (residues 341–345) as calculated from the 60ns high-temperature MD trajectories of RNMT W178C A417C. The color scale represents the number of snapshots in each ‘bin’. (E) Representative snapshots corresponding to the most sampled configuration in panel D. Helix A is shown in blue, hinge in green and lobe in orange. High-transparency image of helix A in the background represents the position in RNMT–RAM crystal structure. (F) MD modelling of the first nucleotide binding mode on the model of best fit obtained from densities by Fabrega et al. (15) (PDB: 1RI2). The predicted direction of the RNA and RAM RNA binding domain (residues 45–90) are indicated. Values in panels C and D represent mean ± S.D. (n = 3).