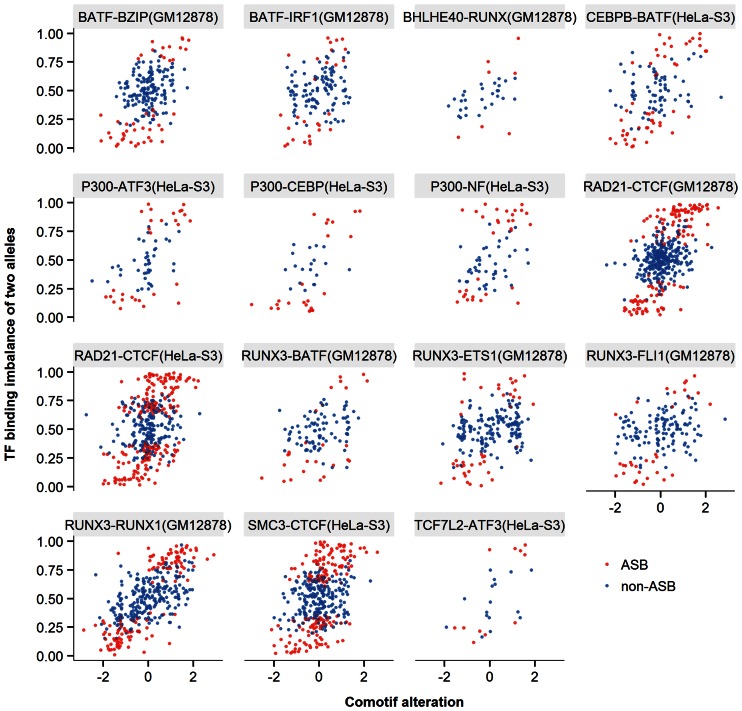
Figure 3.
Alteration of comotif correlated with TF allelic imbalance. The name of each panel specified the ChIP'ed TF followed by the comotif name and the cell line in parentheses. Each dot represented one heterozygous site binding event (red for ASB and blue for non-ASB events) found within the predicted TFBSs of the comotif. The comotif alteration (x-axis) represented the log ratio of motif P-values between the reference and alternative alleles. The allelic binding imbalance (y-axis) indicated the fraction of reads mapped on the reference allele over the whole read coverage at that position. We tested the correlation between the two properties for each ChIP'ed TF and its enriched HOMER motifs, and only significantly correlated pairs were plotted (FDR < 0.05).