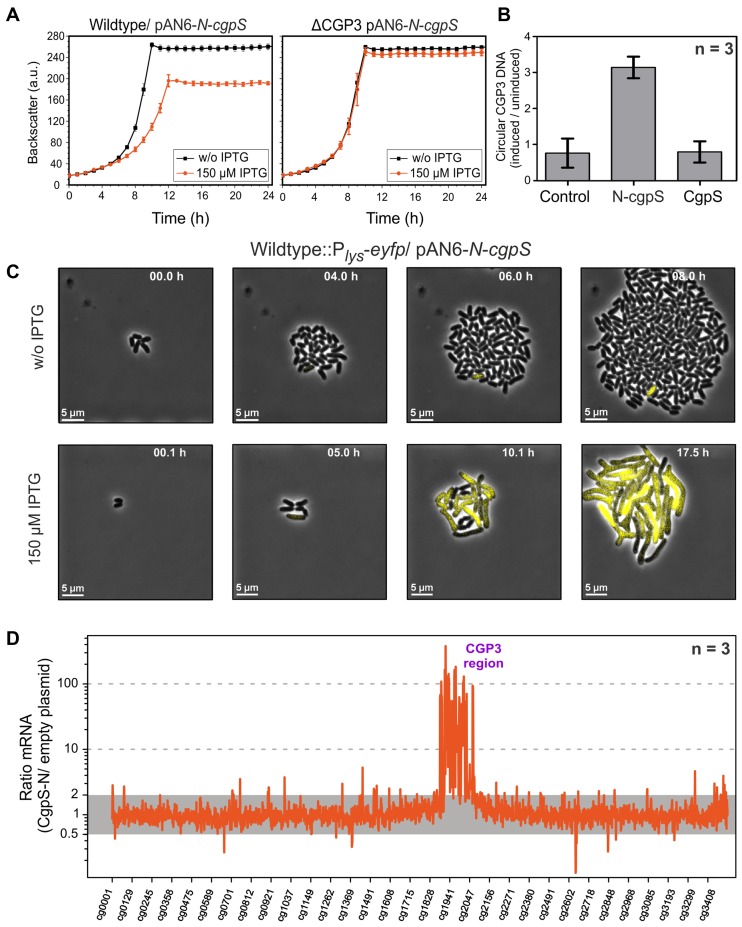
Figure 5.
Countersilencing by overexpression of the N-terminal oligomerization domain of CgpS. (A) Growth studies of the wild-type ATCC 13032 and a strain lacking the CGP3 prophage (ΔCGP3), both carrying the pAN6-N-cgpS overexpression plasmid. Strains were grown in CGXII minimal medium with and without IPTG (150 μM). Data represent average values and standard deviations of three biological replicates. (B) Relative quantification of CGP3 excision using qPCR (20). The N-terminal domain of CgpS and the full-length protein were overproduced as described in (A). Samples for qPCR analysis were taken after 24 h. The relative amounts of circular phage DNA of induced and uninduced samples were compared. As a control wild-type cells with the empty plasmid were used. Data represent average values and standard deviations of three biological replicates. (C) Time-lapse fluorescence microscopy of the C. glutamicum prophage reporter strain (ATCC 13032::Plys-eyfp) carrying the pAN6-N-cgpS. Cells were grown in PDMS-based microfluidic chip devices under continuous supply of CGXII with 25 μg·ml−1 kanamycin and with or without 150 μM IPTG to induce the expression of the truncated CpgS variant (36) (300 nl·min−1) (Video S1 and S2). (D) Comparative transcriptome analysis of C. glutamicum ATCC 13032 containing the overexpression plasmid pAN6-N-cgpS and a strain containing the empty vector control was performed as described in the Materials and Methods section.