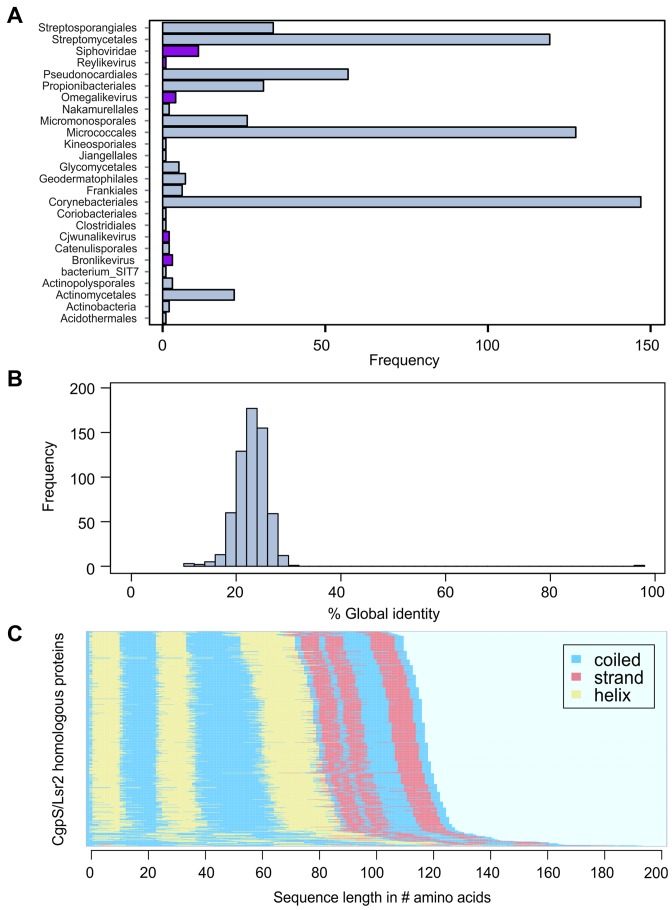
Figure 7.
Orthologous sequences of the CgpS protein in actinomycetes and their phages. (A) Bar chart depicting the frequency of orthologous hits (n = 618) as predicted by PSI-BLAST (39) (e-value ≤ 0.005) across several orders of the phylum Actinobacteria and phages as annotated in the NCBI database (http://www.ncbi.nlm.nih.gov/). Orthologous found in phages are highlighted in purple. (B) Histogram of pairwise global identities between the CgpS amino acid sequence and its orthologous counterparts. The distribution reveals an overall low similarity (mean x = 23.07 and standard deviation σ = 4.05) to the orthologous sequences. Global identity was calculated using the Needleman–Wunsch (40) algorithm from the EMBOSS package (41). (C) Secondary structure prediction calculated by psipred (42) shows conserved protein structure for CgpS and the orthologous amino acid sequences. The corresponding secondary structure of each sequence was ordered in the direction from C-Terminus to N-terminus. Predicted coiled structures are shown in blue, strand regions in red and helices are colored in yellow.